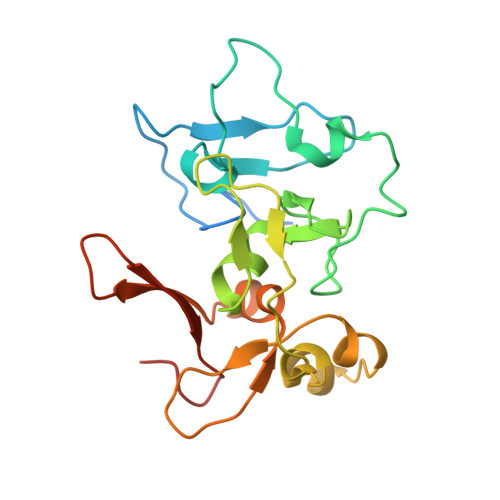
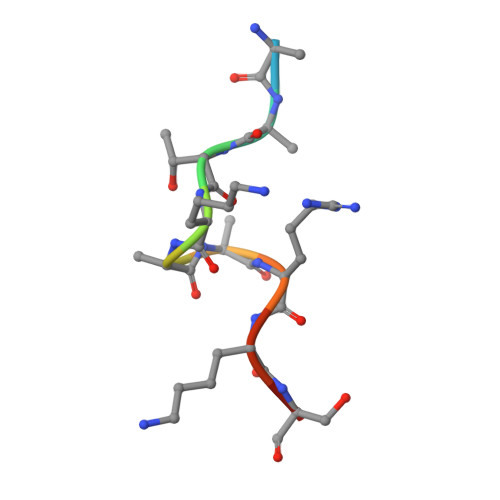
The PZP Domain of AF10 Senses Unmodified H3K27 to Regulate DOT1L-Mediated Methylation of H3K79.
Chen, S., Yang, Z., Wilkinson, A.W., Deshpande, A.J., Sidoli, S., Krajewski, K., Strahl, B.D., Garcia, B.A., Armstrong, S.A., Patel, D.J., Gozani, O.(2015) Mol Cell 60: 319-327
- PubMed: 26439302
- DOI: https://doi.org/10.1016/j.molcel.2015.08.019
- Primary Citation of Related Structures:
5DAG, 5DAH - PubMed Abstract:
AF10, a DOT1L cofactor, is required for H3K79 methylation and cooperates with DOT1L in leukemogenesis. However, the molecular mechanism by which AF10 regulates DOT1L-mediated H3K79 methylation is not clear. Here we report that AF10 contains a "reader" domain that couples unmodified H3K27 recognition to H3K79 methylation. An AF10 region consisting of a PHD finger-Zn knuckle-PHD finger (PZP) folds into a single module that recognizes amino acids 22-27 of H3, and this interaction is abrogated by H3K27 modification. Structural studies reveal that H3 binding triggers rearrangement of the PZP module to form an H3(22-27)-accommodating channel and that the unmodified H3K27 side chain is encased in a compact hydrogen-bond acceptor-lined cage. In cells, PZP recognition of H3 is required for H3K79 dimethylation, expression of DOT1L-target genes, and proliferation of DOT1L-addicted leukemic cells. Together, our results uncover a pivotal role for H3K27-via readout by the AF10 PZP domain-in regulating the cancer-associated enzyme DOT1L.
- Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: