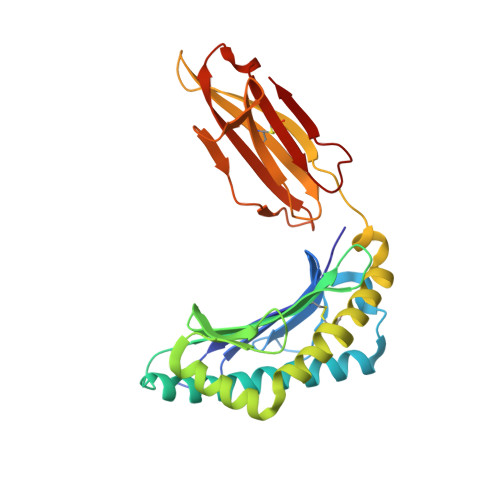
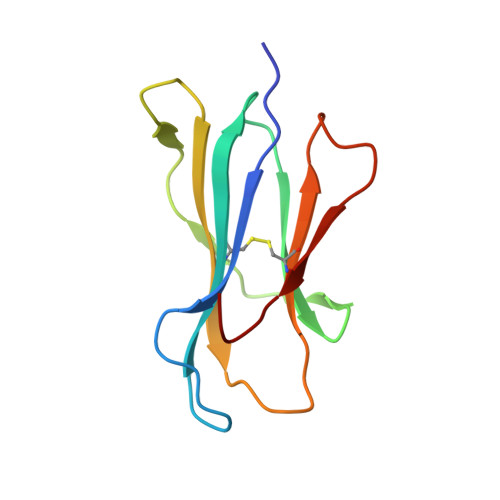
Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove.
McMurtrey, C., Trolle, T., Sansom, T., Remesh, S.G., Kaever, T., Bardet, W., Jackson, K., McLeod, R., Sette, A., Nielsen, M., Zajonc, D.M., Blader, I.J., Peters, B., Hildebrand, W.(2016) Elife 5
- PubMed: 26824387
- DOI: https://doi.org/10.7554/eLife.12556
- Primary Citation of Related Structures:
5D9S, 5DDH - PubMed Abstract:
HLA class I presentation of pathogen-derived peptide ligands is essential for CD8+ T-cell recognition of Toxoplasma gondii infected cells. Currently, little data exist pertaining to peptides that are presented after T. gondii infection. Herein we purify HLA-A*02:01 complexes from T. gondii infected cells and characterize the peptide ligands using LCMS. We identify 195 T. gondii encoded ligands originating from both secreted and cytoplasmic proteins. Surprisingly, T. gondii ligands are significantly longer than uninfected host ligands, and these longer pathogen-derived peptides maintain a canonical N-terminal binding core yet exhibit a C-terminal extension of 1-30 amino acids. Structural analysis demonstrates that binding of extended peptides opens the HLA class I F' pocket, allowing the C-terminal extension to protrude through one end of the binding groove. In summary, we demonstrate that unrealized structural flexibility makes MHC class I receptive to parasite-derived ligands that exhibit unique C-terminal peptide extensions.
- Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center, Oklahoma City, United States.
Organizational Affiliation: