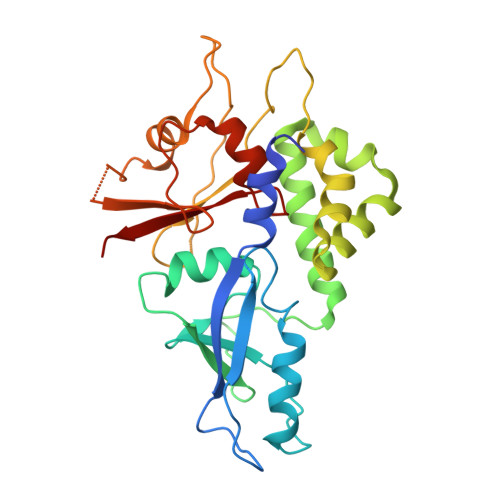
Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay.
Kim, M., Sandford, E., Gatica, D., Qiu, Y., Liu, X., Zheng, Y., Schulman, B.A., Xu, J., Semple, I., Ro, S.H., Kim, B., Mavioglu, R.N., Tolun, A., Jipa, A., Takats, S., Karpati, M., Li, J.Z., Yapici, Z., Juhasz, G., Lee, J.H., Klionsky, D.J., Burmeister, M.(2016) Elife 5
- PubMed: 26812546
- DOI: https://doi.org/10.7554/eLife.12245
- Primary Citation of Related Structures:
5D7G - PubMed Abstract:
Autophagy is required for the homeostasis of cellular material and is proposed to be involved in many aspects of health. Defects in the autophagy pathway have been observed in neurodegenerative disorders; however, no genetically-inherited pathogenic mutations in any of the core autophagy-related (ATG) genes have been reported in human patients to date. We identified a homozygous missense mutation, changing a conserved amino acid, in ATG5 in two siblings with congenital ataxia, mental retardation, and developmental delay. The subjects' cells display a decrease in autophagy flux and defects in conjugation of ATG12 to ATG5. The homologous mutation in yeast demonstrates a 30-50% reduction of induced autophagy. Flies in which Atg5 is substituted with the mutant human ATG5 exhibit severe movement disorder, in contrast to flies expressing the wild-type human protein. Our results demonstrate the critical role of autophagy in preventing neurological diseases and maintaining neuronal health.
- Department of Molecular and Integrative Physiology, University of Michigan, Ann Arbor, United States.
Organizational Affiliation: