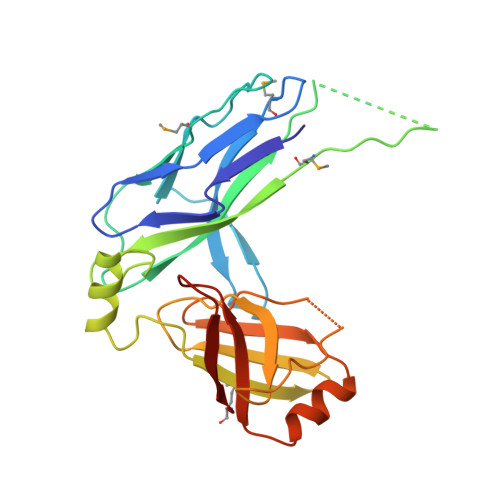
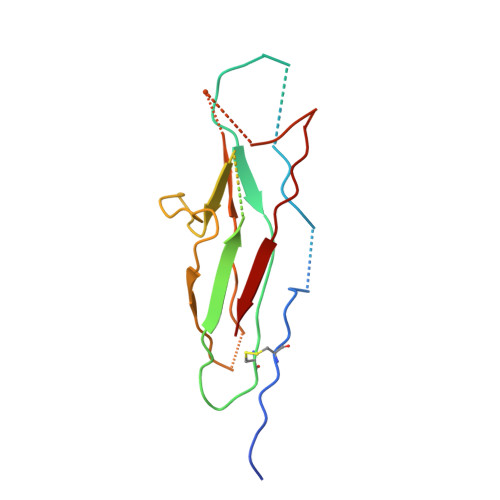
Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Pakharukova, N., Garnett, J.A., Tuittila, M., Paavilainen, S., Diallo, M., Xu, Y., Matthews, S.J., Zavialov, A.V.(2015) PLoS Pathog 11: e1005269-e1005269
- PubMed: 26587649
- DOI: https://doi.org/10.1371/journal.ppat.1005269
- Primary Citation of Related Structures:
5D6H, 5DFK - PubMed Abstract:
Gram-negative pathogens express fibrous adhesive organelles that mediate targeting to sites of infection. The major class of these organelles is assembled via the classical, alternative and archaic chaperone-usher pathways. Although non-classical systems share a wider phylogenetic distribution and are associated with a range of diseases, little is known about their assembly mechanisms. Here we report atomic-resolution insight into the structure and biogenesis of Acinetobacter baumannii Csu and Escherichia coli ECP biofilm-mediating pili. We show that the two non-classical systems are structurally related, but their assembly mechanism is strikingly different from the classical assembly pathway. Non-classical chaperones, unlike their classical counterparts, maintain subunits in a substantially disordered conformational state, akin to a molten globule. This is achieved by a unique binding mechanism involving the register-shifted donor strand complementation and a different subunit carboxylate anchor. The subunit lacks the classical pre-folded initiation site for donor strand exchange, suggesting that recognition of its exposed hydrophobic core starts the assembly process and provides fresh inspiration for the design of inhibitors targeting chaperone-usher systems.
- Department of Chemistry, University of Turku, Turku, JBL, Arcanum, Turku, Finland.
Organizational Affiliation: