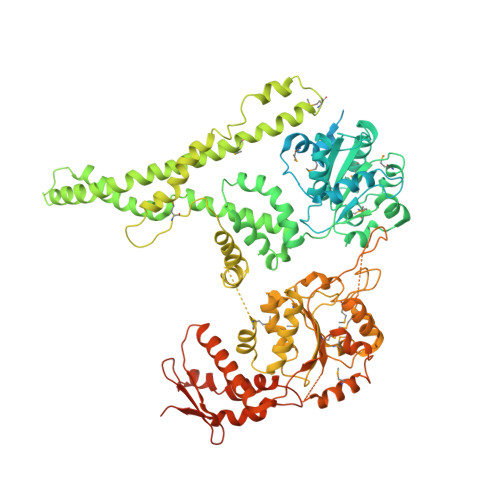
Structural basis for the disaggregase activity and regulation of Hsp104.
Heuck, A., Schitter-Sollner, S., Suskiewicz, M.J., Kurzbauer, R., Kley, J., Schleiffer, A., Rombaut, P., Herzog, F., Clausen, T.(2016) Elife 5
- PubMed: 27901467
- DOI: https://doi.org/10.7554/eLife.21516
- Primary Citation of Related Structures:
5D4W - PubMed Abstract:
The Hsp104 disaggregase is a two-ring ATPase machine that rescues various forms of non-native proteins including the highly resistant amyloid fibers. The structural-mechanistic underpinnings of how the recovery of toxic protein aggregates is promoted and how this potent unfolding activity is prevented from doing collateral damage to cellular proteins are not well understood. Here, we present structural and biochemical data revealing the organization of Hsp104 from Chaetomium thermophilum at 3.7 Å resolution. We show that the coiled-coil domains encircling the disaggregase constitute a 'restraint mask' that sterically controls the mobility and thus the unfolding activity of the ATPase modules. In addition, we identify a mechanical linkage that coordinates the activity of the two ATPase rings and accounts for the high unfolding potential of Hsp104. Based on these findings, we propose a general model for how Hsp104 and related chaperones operate and are kept under control until recruited to appropriate substrates.
- Research Institute of Molecular Pathology, Vienna Biocenter, Vienna, Austria.
Organizational Affiliation: