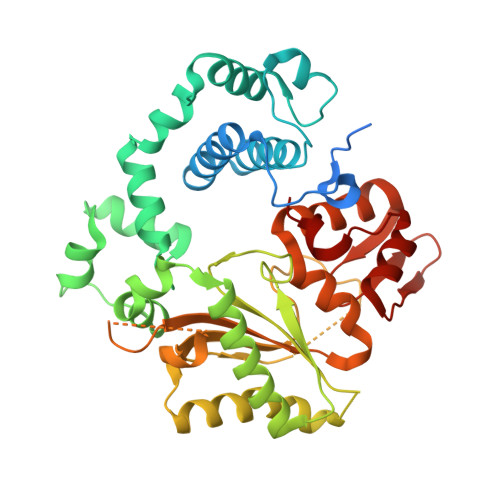
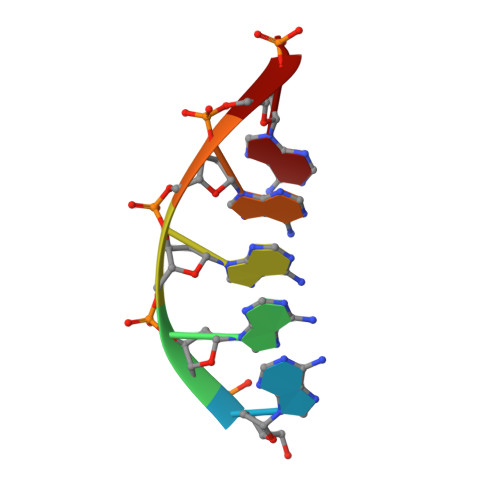
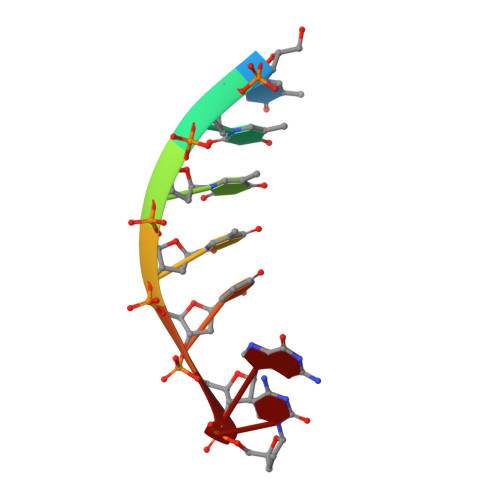
Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination.
Loc'h, J., Rosario, S., Delarue, M.(2016) Structure 24: 1452-1463
- PubMed: 27499438
- DOI: https://doi.org/10.1016/j.str.2016.06.014
- Primary Citation of Related Structures:
5D46, 5D49, 5D4B - PubMed Abstract:
Eukaryotic DNA polymerase of the polX family, such as pol μ and terminal deoxynucleotidyl transferase (TdT), are key components of the non-homologous end-joining or V(D)J recombination machinery, respectively. The established role of TdT is to add random nucleotides during V(D)J recombination. Here we show that TdT also has a templated-polymerase activity, similar to pol μ, in the presence of higher concentrations of a downstream DNA duplex, and performs a micro-homology single base-pair search to align the DNA synapsis. To understand the molecular basis of this alignment, we solve crystal structures of TdT with four DNA strands and study the influence of the 3' protruding end. Two mutations in TdT inspired by sequence alignments with pol μ further improve the templated activity. We propose that both templated and untemplated activities of TdT are needed to explain the distributions of lengths of N regions observed experimentally in T cell receptors and antibodies.
- Unité de Dynamique Structurale des Macromolécules, Institut Pasteur, UMR 3528 du C.N.R.S., 25 rue du Dr Roux, 75015 Paris, France.
Organizational Affiliation: