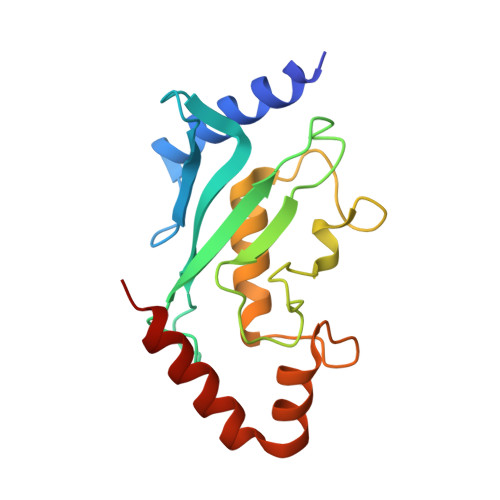
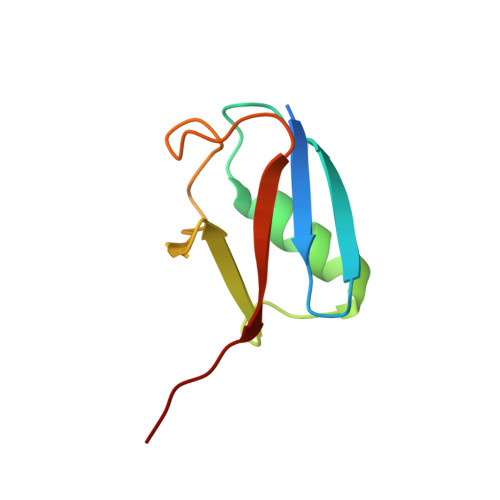
Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Cappadocia, L., Pichler, A., Lima, C.D.(2015) Nat Struct Mol Biol 22: 968-975
- PubMed: 26524494
- DOI: https://doi.org/10.1038/nsmb.3116
- Primary Citation of Related Structures:
5D2M - PubMed Abstract:
E3 protein ligases enhance transfer of ubiquitin-like (Ubl) proteins from E2 conjugating enzymes to substrates by stabilizing the thioester-charged E2~Ubl in a closed configuration optimally aligned for nucleophilic attack. Here, we report biochemical and structural data that define the N-terminal domain of the Homo sapiens ZNF451 as the catalytic module for SUMO E3 ligase activity. The ZNF451 catalytic module contains tandem SUMO-interaction motifs (SIMs) bridged by a Pro-Leu-Arg-Pro (PLRP) motif. The first SIM and PLRP motif engage thioester-charged E2~SUMO while the next SIM binds a second molecule of SUMO bound to the back side of E2. We show that ZNF451 is SUMO2 specific and that SUMO modification of ZNF451 may contribute to activity by providing a second molecule of SUMO that interacts with E2. Our results are consistent with ZNF451 functioning as a bona fide SUMO E3 ligase.
- Structural Biology Program, Sloan Kettering Institute, New York, New York, USA.
Organizational Affiliation: