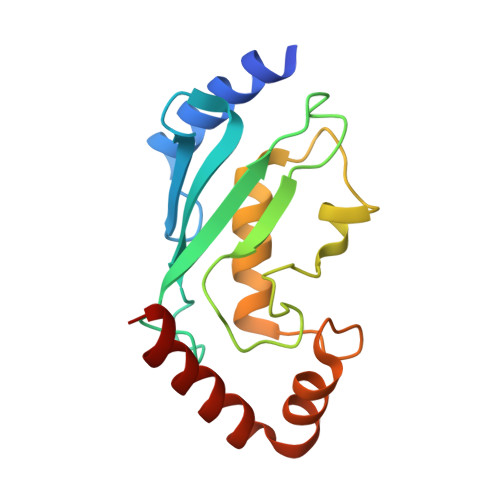
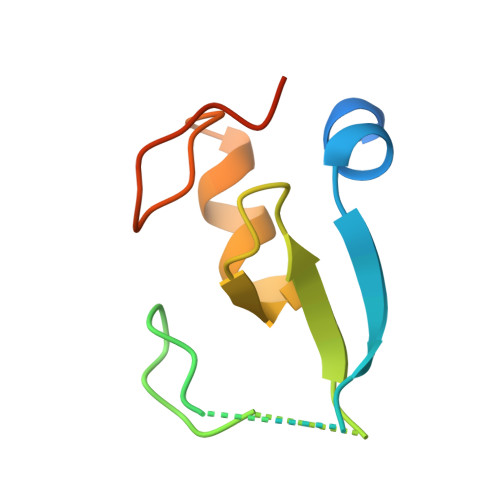
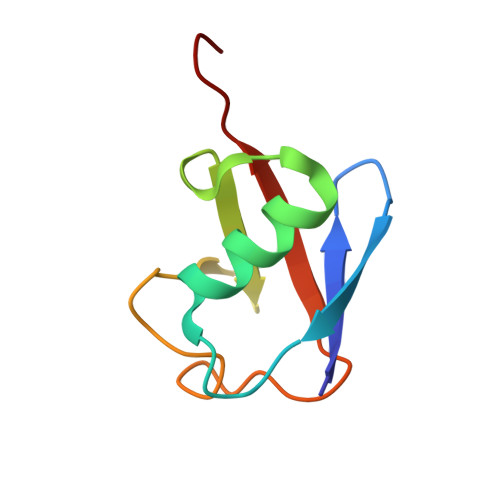
Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Wright, J.D., Mace, P.D., Day, C.L.(2016) Nat Struct Mol Biol 23: 45-52
- PubMed: 26656854
- DOI: https://doi.org/10.1038/nsmb.3142
- Primary Citation of Related Structures:
5D0I, 5D0K, 5D0M - PubMed Abstract:
RING-domain E3 ligases enhance transfer of ubiquitin to substrate proteins by stabilizing the RING-bound thioester-linked E2∼ubiquitin conjugate in a defined conformation that primes the active site for nucleophilic attack. Here we report that the monomeric RING domains from the human E3 ligases Arkadia and Ark2C bind directly to free ubiquitin with an affinity comparable to that of other dedicated ubiquitin-binding domains. Further work showed that the Ark-like RING domain and the noncovalently bound ubiquitin molecule coordinately stabilize the E2-conjugated ubiquitin (donor ubiquitin) in the 'closed' conformation. Our studies identify the RING domain of Arkadia as a ubiquitin-binding domain and provide insight into a new ubiquitin-dependent mechanism used by monomeric RING domains to activate ubiquitin transfer. This study also suggests how substrates that have been monoubiquitinated could be favored for further ubiquitination.
- Biochemistry Department, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: