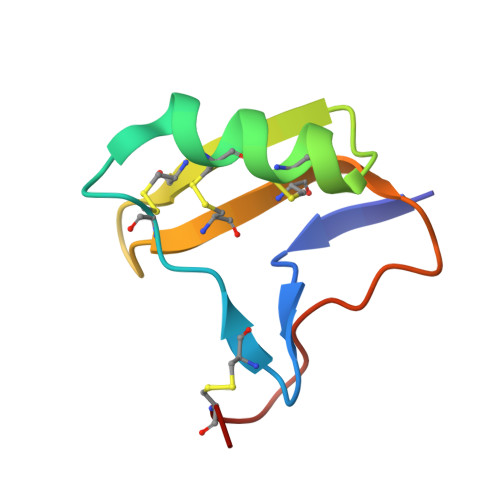
Elucidation of the Covalent and Tertiary Structures of Biologically Active Ts3 Toxin.
Dang, B., Kubota, T., Mandal, K., Correa, A.M., Bezanilla, F., Kent, S.B.(2016) Angew Chem Int Ed Engl 55: 8639-8642
- PubMed: 27244051
- DOI: https://doi.org/10.1002/anie.201603420
- Primary Citation of Related Structures:
5CY0 - PubMed Abstract:
Ts3 is an alpha scorpion toxin from the venom of the Brazilian scorpion Tityus serrulatus. Ts3 binds to the domain IV voltage sensor of voltage-gated sodium channels (Nav ) and slows down their fast inactivation. The covalent structure of the Ts3 toxin is uncertain, and the structure of the folded protein molecule is unknown. Herein, we report the total chemical synthesis of four candidate Ts3 toxin protein molecules and the results of structure-activity studies that enabled us to establish the covalent structure of biologically active Ts3 toxin. We also report the synthesis of the mirror image form of the Ts3 protein molecule, and the use of racemic protein crystallography to determine the folded (tertiary) structure of biologically active Ts3 toxin by X-ray diffraction.
- Department of Chemistry, University of Chicago, Chicago, IL, 60637, USA.
Organizational Affiliation: