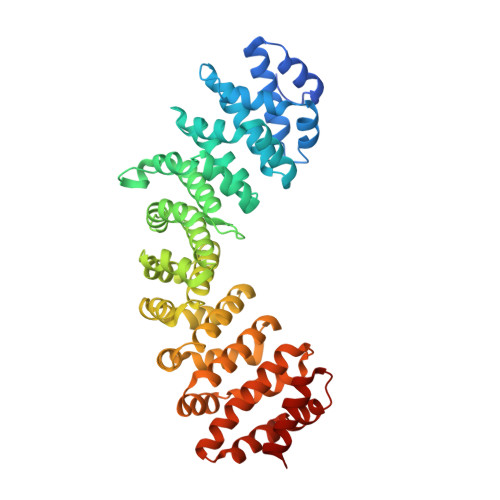
Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3
Park, J.K., Das, T., Song, E.J., Kim, E.E.(2016) Nucleic Acids Res 44: 5424-5437
- PubMed: 27060135
- DOI: https://doi.org/10.1093/nar/gkw218
- Primary Citation of Related Structures:
5CTQ, 5CTR, 5CTT - PubMed Abstract:
Squamous cell carcinoma antigen recognized by T-cells 3 (SART3) is a U4/U6 recycling factor as well as a targeting factor of USP4 and USP15. However, the details of how SART3 recognizes these deubiquitinases and how they get subsequently translocated into the nucleus are not known. Here, we present the crystal structures of the SART3 half-a-tetratricopeptide (HAT) repeat domain alone and in complex with the domain present in ubiquitin-specific protease (DUSP)-ubiquitin-like (UBL) domains of ubiquitin specific protease 4 (USP4). The 12 HAT repeats of SART3 are in two sub-domains (HAT-N and HAT-C) forming a dimer through HAT-C. USP4 binds SART3 at the opposite surface of the HAT-C dimer interface utilizing the β-structured linker between the DUSP and the UBL domains. The binding affinities of USP4 and USP15 to SART3 are 0.9 μM and 0.2 μM, respectively. The complex structure of SART3 nuclear localization signal (NLS) and importin-α reveals bipartite binding, and removal of SART3 NLS prevents the entry of USP4 (and USP15) into the nucleus and abrogates the subsequent deubiquitinase activity of USP4.
- Biomedical Research Institute, Korea Institute of Science and Technology, Hwarangno 14-gil 5, Seongbuk-gu, Seoul 136-791, Republic of Korea.
Organizational Affiliation: