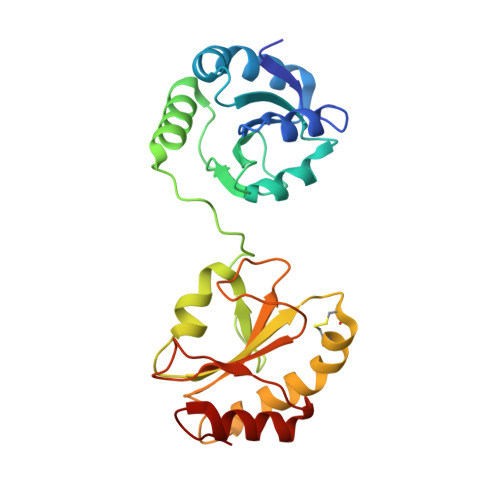
Structural basis of redox-dependent substrate binding of protein disulfide isomerase.
Yagi-Utsumi, M., Satoh, T., Kato, K.(2015) Sci Rep 5: 13909-13909
- PubMed: 26350503
- DOI: https://doi.org/10.1038/srep13909
- Primary Citation of Related Structures:
5CRW - PubMed Abstract:
Protein disulfide isomerase (PDI) is a multidomain enzyme, operating as an essential folding catalyst, in which the b' and a' domains provide substrate binding sites and undergo an open-closed domain rearrangement depending on the redox states of the a' domain. Despite the long research history of this enzyme, three-dimensional structural data remain unavailable for its ligand-binding mode. Here we characterize PDI substrate recognition using α-synuclein (αSN) as the model ligand. Our nuclear magnetic resonance (NMR) data revealed that the substrate-binding domains of PDI captured the αSN segment Val37-Val40 only in the oxidized form. Furthermore, we determined the crystal structure of an oxidized form of the b'-a' domains in complex with an undecapeptide corresponding to this segment. The peptide-binding mode observed in the crystal structure with NMR validation, was characterized by hydrophobic interactions on the b' domain in an open conformation. Comparison with the previously reported crystal structure indicates that the a' domain partially masks the binding surface of the b' domain, causing steric hindrance against the peptide in the reduced form of the b'-a' domains that exhibits a closed conformation. These findings provide a structural basis for the mechanism underlying the redox-dependent substrate binding of PDI.
- Okazaki Institute for Integrative Bioscience and Institute for Molecular Science, National Institutes of Natural Sciences, 5-1 Higashiyama, Myodaiji, Okazaki, Aichi 444-8787, Japan.
Organizational Affiliation: