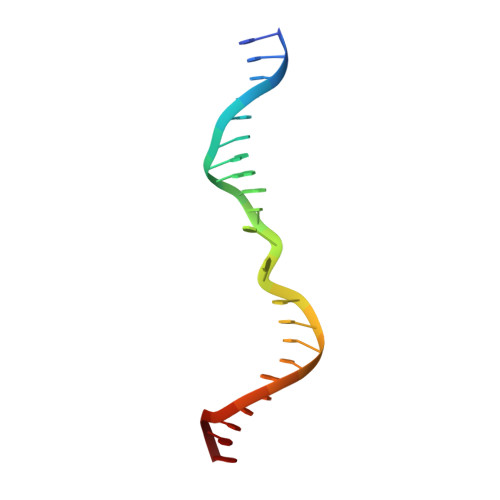
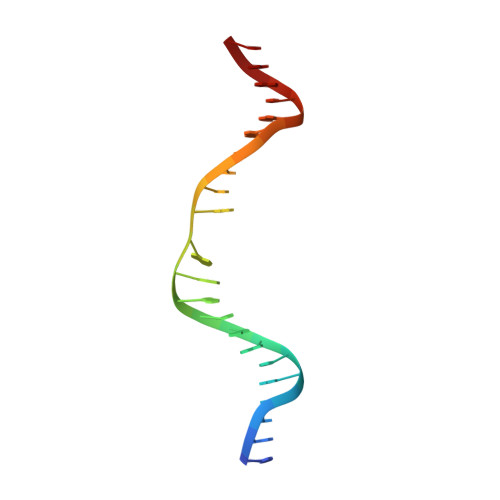
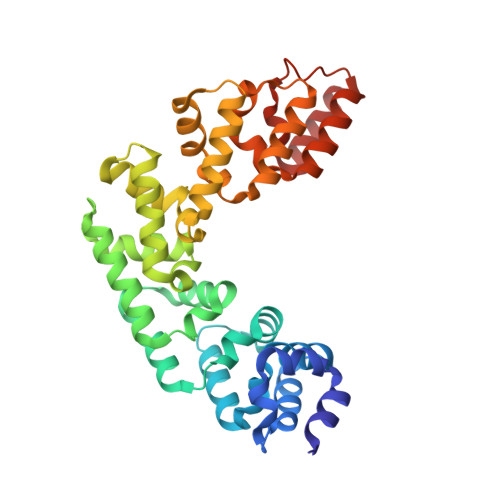Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
Byrnes, J., Hauser, K., Norona, L., Mejia, E., Simmerling, C., Garcia-Diaz, M.(2016) J Mol Biology 428: 2542-2556
- PubMed: 26523681
- DOI: https://doi.org/10.1016/j.jmb.2015.10.021
- Primary Citation of Related Structures:
5CKY, 5CO0, 5CRJ, 5CRK - PubMed Abstract:
Human mitochondrial transcription termination occurs within the leu-tRNA gene and is mediated by the DNA binding protein MTERF1. The crystal structure of MTERF1 bound to the canonical termination sequence reveals a rare base flipping event that involves the eversion of three nucleotides. These nucleotides are stabilized by stacking interactions with three MTERF1 residues, which are essential not only for base flipping but also for termination activity. To further understand the mechanism of base flipping, we examined each of the individual stacking interactions in structural, energetic and functional detail. Individual substitutions of Arg162, Tyr288 and Phe243 have revealed unequal contributions to overall termination activity. Furthermore, our work identifies an important role for Phe322 in the base flipping mechanism and we demonstrate how Phe322 and Phe243 are important for coupling base flipping between the heavy and light strand DNA chains. We propose a stepwise model for the base flipping process that recapitulates our observations. Finally, we show that MTERF1 has the ability to accommodate alternate active conformations. The adaptability of base flipping has implications for MTERF1 function and for the putative function of MTERF1 at alternative binding sites in human mitochondria.
- Department of Pharmacological Sciences, Stony Brook University, Stony Brook, NY 11794, USA.
Organizational Affiliation: