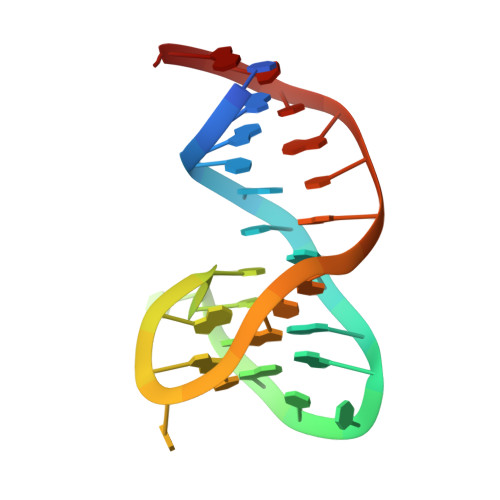
Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding.
Russo Krauss, I., Spiridonova, V., Pica, A., Napolitano, V., Sica, F.(2016) Nucleic Acids Res 44: 983-991
- PubMed: 26673709
- DOI: https://doi.org/10.1093/nar/gkv1384
- Primary Citation of Related Structures:
5CMX - PubMed Abstract:
Mixed duplex/quadruplex oligonucleotides have attracted great interest as therapeutic targets as well as effective biomedical aptamers. In the case of thrombin-binding aptamer (TBA), the addition of a duplex motif to the G-quadruplex module improves the aptamer resistance to biodegradation and the affinity for thrombin. In particular, the mixed oligonucleotide RE31 is significantly more effective than TBA in anticoagulation experiments and shows a slower disappearance rate in human plasma and blood. In the crystal structure of the complex with thrombin, RE31 adopts an elongated structure in which the duplex and quadruplex regions are perfectly stacked on top of each other, firmly connected by a well-structured junction. The lock-and-key shape complementarity between the TT loops of the G-quadruplex and the protein exosite I gives rise to the basic interaction that stabilizes the complex. However, our data suggest that the duplex motif may have an active role in determining the greater anti-thrombin activity in biological fluids with respect to TBA. This work gives new information on mixed oligonucleotides and highlights the importance of structural data on duplex/quadruplex junctions, which appear to be varied, unpredictable, and fundamental in determining the aptamer functional properties.
- Department of Chemical Sciences, University of Naples 'Federico II', Naples, Italy Institute of Biostructures and Bioimages, C.N.R, Naples, Italy.
Organizational Affiliation: