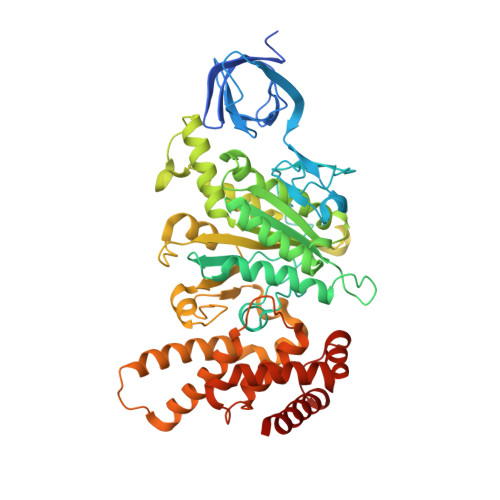
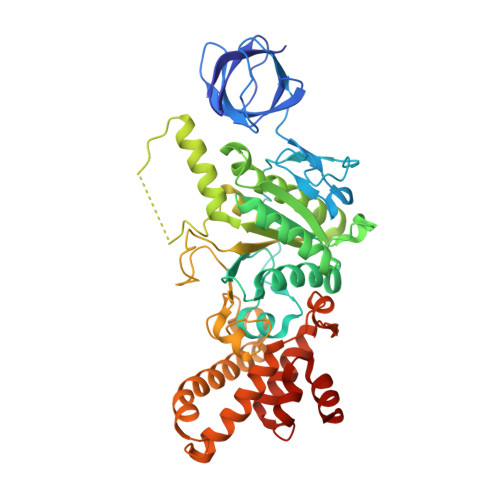
Structure of a catalytic dimer of the alpha- and beta-subunits of the F-ATPase from Paracoccus denitrificans at 2.3 angstrom resolution.
Morales-Rios, E., Montgomery, M.G., Leslie, A.G., Garcia-Trejo, J.J., Walker, J.E.(2015) Acta Crystallogr F Struct Biol Commun 71: 1309-1317
- PubMed: 26457523
- DOI: https://doi.org/10.1107/S2053230X15016076
- Primary Citation of Related Structures:
5CDF - PubMed Abstract:
The structures of F-ATPases have predominantly been determined from mitochondrial enzymes, and those of the enzymes in eubacteria have been less studied. Paracoccus denitrificans is a member of the α-proteobacteria and is related to the extinct protomitochondrion that became engulfed by the ancestor of eukaryotic cells. The P. denitrificans F-ATPase is an example of a eubacterial F-ATPase that can carry out ATP synthesis only, whereas many others can catalyse both the synthesis and the hydrolysis of ATP. Inhibition of the ATP hydrolytic activity of the P. denitrificans F-ATPase involves the ζ inhibitor protein, an α-helical protein that binds to the catalytic F1 domain of the enzyme. This domain is a complex of three α-subunits and three β-subunits, and one copy of each of the γ-, δ- and ℇ-subunits. Attempts to crystallize the F1-ζ inhibitor complex yielded crystals of a subcomplex of the catalytic domain containing the α- and β-subunits only. Its structure was determined to 2.3 Å resolution and consists of a heterodimer of one α-subunit and one β-subunit. It has no bound nucleotides, and it corresponds to the `open' or `empty' catalytic interface found in other F-ATPases. The main significance of this structure is that it aids in the determination of the structure of the intact membrane-bound F-ATPase, which has been crystallized.
- The Medical Research Council Mitochondrial Biology Unit, Cambridge Biomedical Campus, Hills Road, Cambridge CB2 0XY, England.
Organizational Affiliation: