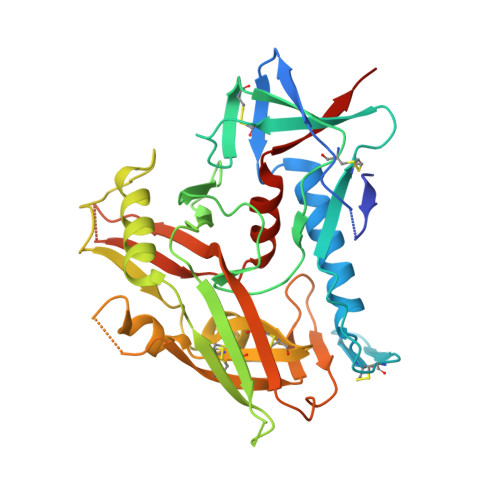
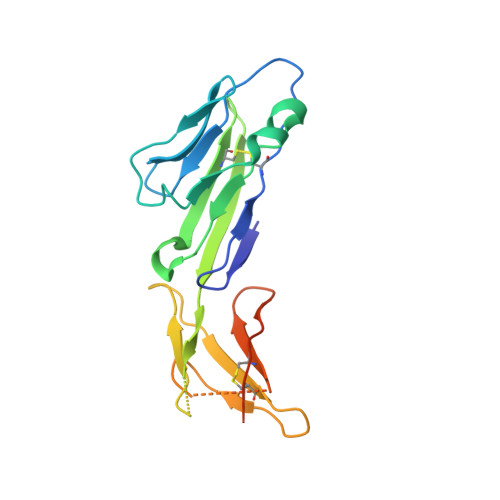
Structure of an HIV-2 gp120 in Complex with CD4.
Davenport, Y.W., West, A.P., Bjorkman, P.J.(2015) J Virol 90: 2112-2118
- PubMed: 26608312
- DOI: https://doi.org/10.1128/JVI.02678-15
- Primary Citation of Related Structures:
5CAY - PubMed Abstract:
HIV-2 is a nonpandemic form of the virus causing AIDS, and the majority of HIV-2-infected patients exhibit long-term nonprogression. The HIV-1 and HIV-2 envelope glycoproteins, the sole targets of neutralizing antibodies, share 30 to 40% identity. As a first step in understanding the reduced pathogenicity of HIV-2, we solved a 3.0-Å structure of an HIV-2 gp120 bound to the host receptor CD4, which reveals structural similarity to HIV-1 gp120 despite divergence in amino acid sequence.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, California, USA.
Organizational Affiliation: