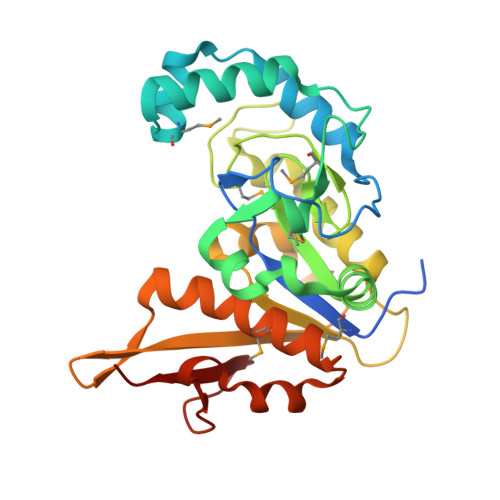
The DUF328 family member YaaA is a DNA-binding protein with a novel fold.
Prahlad, J., Yuan, Y., Lin, J., Chang, C.W., Iwata-Reuyl, D., Liu, Y., de Crecy-Lagard, V., Wilson, M.A.(2020) J Biological Chem 295: 14236-14247
- PubMed: 32796037
- DOI: https://doi.org/10.1074/jbc.RA120.015055
- Primary Citation of Related Structures:
5CAJ - PubMed Abstract:
DUF328 family proteins are present in many prokaryotes; however, their molecular activities are unknown. The Escherichia coli DUF328 protein YaaA is a member of the OxyR regulon and is protective against oxidative stress. Because uncharacterized proteins involved in prokaryotic oxidative stress response are rare, we sought to learn more about the DUF328 family. Using comparative genomics, we found a robust association between the DUF328 family and genes involved in DNA recombination and the oxidative stress response. In some proteins, DUF328 domains are fused to other domains involved in DNA binding, recombination, and repair. Cofitness analysis indicates that DUF328 family genes associate with recombination-mediated DNA repair pathways, particularly the RecFOR pathway. Purified recombinant YaaA binds to dsDNA, duplex DNA containing bubbles of unpaired nucleotides, and Holliday junction constructs in vitro with dissociation equilibrium constants of 200-300 nm YaaA binds DNA with positive cooperativity, forming multiple shifted species in electrophoretic mobility shift assays. The 1.65-Å resolution X-ray crystal structure of YaaA reveals that the protein possesses a new fold that we name the cantaloupe fold. YaaA has a positively charged cleft and a helix-hairpin-helix DNA-binding motif found in other DNA repair enzymes. Our results demonstrate that YaaA is a new type of DNA-binding protein associated with the oxidative stress response and that this molecular function is likely conserved in other DUF328 family members.
- Department of Biochemistry and Redox Biology Center, University of Nebraska, Lincoln, Nebraska, USA.
Organizational Affiliation: