A YidC-like Protein in the Archaeal Plasma Membrane.
Borowska, M.T., Dominik, P.K., Anghel, S.A., Kossiakoff, A.A., Keenan, R.J.(2015) Structure 23: 1715-1724
- PubMed: 26256539
- DOI: https://doi.org/10.1016/j.str.2015.06.025
- Primary Citation of Related Structures:
5C8J - PubMed Abstract:
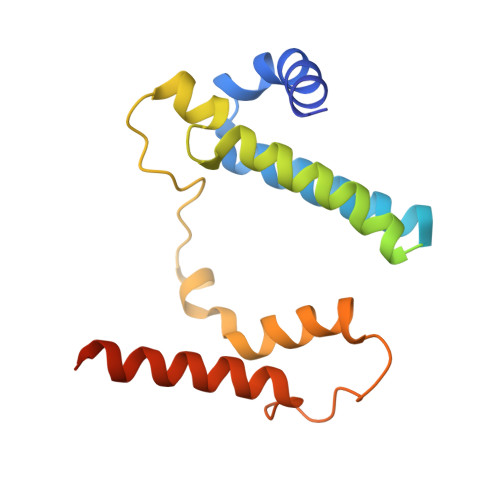
Cells possess specialized machinery to direct the insertion of membrane proteins into the lipid bilayer. In bacteria, the essential protein YidC inserts certain proteins into the plasma membrane, and eukaryotic orthologs are present in the mitochondrial inner membrane and the chloroplast thylakoid membrane. The existence of homologous insertases in archaea has been proposed based on phylogenetic analysis. However, limited sequence identity, distinct architecture, and the absence of experimental data have made this assignment ambiguous. Here we describe the 3.5-Å crystal structure of an archaeal DUF106 protein from Methanocaldococcus jannaschii (Mj0480), revealing a lipid-exposed hydrophilic surface presented by a conserved YidC-like fold. Functional analysis reveals selective binding of Mj0480 to ribosomes displaying a stalled YidC substrate, and a direct interaction between the buried hydrophilic surface of Mj0480 and the nascent chain. These data provide direct experimental evidence that the archaeal DUF106 proteins are YidC/Oxa1/Alb3-like insertases of the archaeal plasma membrane.
- Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57(th) Street, Chicago, IL 60637, USA.
Organizational Affiliation: