Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.
Kong, L., Torrents de la Pena, A., Deller, M.C., Garces, F., Sliepen, K., Hua, Y., Stanfield, R.L., Sanders, R.W., Wilson, I.A.(2015) Acta Crystallogr D Biol Crystallogr 71: 2099-2108
- PubMed: 26457433
- DOI: https://doi.org/10.1107/S1399004715013917
- Primary Citation of Related Structures:
5C7K - PubMed Abstract:
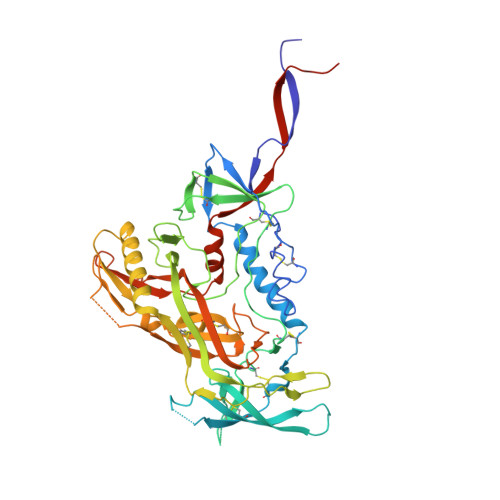
The HIV-1 envelope gp160 glycoprotein (Env) is a trimer of gp120 and gp41 heterodimers that mediates cell entry and is the primary target of the humoral immune response. Broadly neutralizing antibodies (bNAbs) to HIV-1 have revealed multiple epitopes or sites of vulnerability, but mapping of most of these sites is incomplete owing to a paucity of structural information on the full epitope in the context of the Env trimer. Here, a crystal structure of the soluble BG505 SOSIP gp140 trimer at 4.6 Å resolution with the bNAbs 8ANC195 and PGT128 reveals additional interactions in comparison to previous antibody-gp120 structures. For 8ANC195, in addition to previously documented interactions with gp120, a substantial interface with gp41 is now elucidated that includes extensive interactions with the N637 glycan. Surprisingly, removal of the N637 glycan did not impact 8ANC195 affinity, suggesting that the antibody has evolved to accommodate this glycan without loss of binding energy. PGT128 indirectly affects the N262 glycan by a domino effect, in which PGT128 binds to the N301 glycan, which in turn interacts with and repositions the N262 glycan, thereby illustrating the important role of neighboring glycans on epitope conformation and stability. Comparisons with other Env trimer and gp120 structures support an induced conformation for glycan N262, suggesting that the glycan shield is allosterically modified upon PGT128 binding. These complete epitopes of two broadly neutralizing antibodies on the Env trimer can now be exploited for HIV-1 vaccine design.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, USA.
Organizational Affiliation: