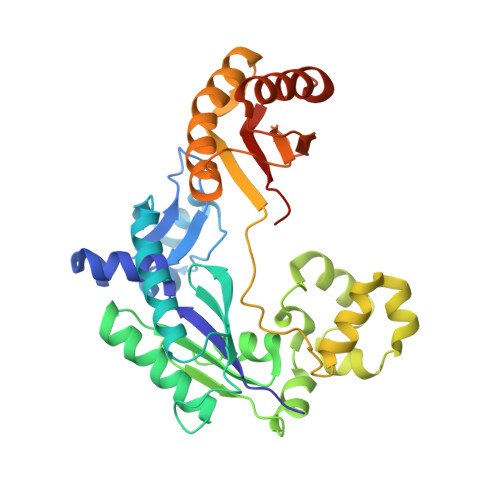
Reactive Oxygen Species Play an Important Role in the Bactericidal Activity of Quinolone Antibiotics
Kottur, J., Nair, D.T.(2016) Angew Chem Int Ed Engl 55: 2397-2400
- PubMed: 26757158
- DOI: https://doi.org/10.1002/anie.201509340
- Primary Citation of Related Structures:
5C5J - PubMed Abstract:
Recent studies posit that reactive oxygen species (ROS) contribute to the cell lethality of bactericidal antibiotics. However, this conjecture has been challenged and remains controversial. To resolve this controversy, we adopted a strategy that involves DNA polymerase IV (PolIV). The nucleotide pool of the cell gets oxidized by ROS and PolIV incorporates the damaged nucleotides (especially 8oxodGTP) into the genome, which results in death of the bacteria. By using a combination of structural and biochemical tools coupled with growth assays, it was shown that selective perturbation of the 8oxodGTP incorporation activity of PolIV results in considerable enhancement of the survival of bacteria in the presence of the norfloxacin antibiotic. Our studies therefore indicate that ROS induced in bacteria by the presence of antibiotics in the environment contribute significantly to cell lethality.
- Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad, 121001, India.
Organizational Affiliation: