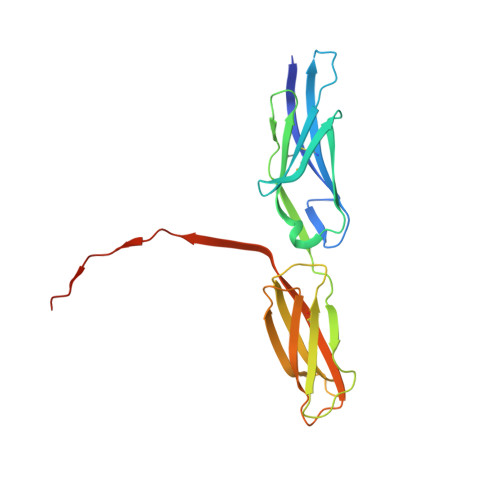
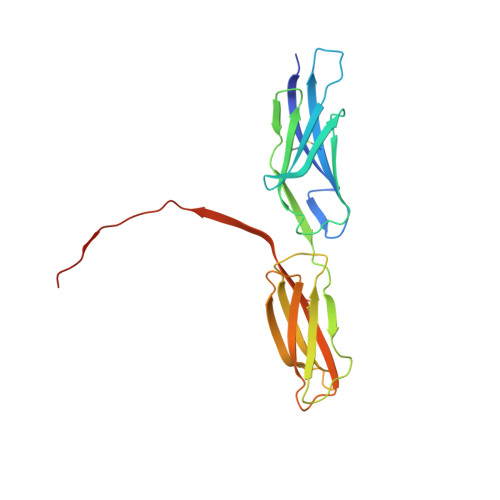
Structural basis for PECAM-1 homophilic binding.
Paddock, C., Zhou, D., Lertkiatmongkol, P., Newman, P.J., Zhu, J.(2016) Blood 127: 1052-1061
- PubMed: 26702061
- DOI: https://doi.org/10.1182/blood-2015-07-660092
- Primary Citation of Related Structures:
5C14 - PubMed Abstract:
Platelet endothelial cell adhesion molecule-1 (PECAM-1) is a 130-kDa member of the immunoglobulin gene superfamily (IgSF) that is present on the surface of circulating platelets and leukocytes, and highly expressed at the junctions of confluent endothelial cell monolayers. PECAM-1-mediated homophilic interactions, known to be mediated by its 2 amino-terminal immunoglobulin homology domains, are essential for concentrating PECAM-1 at endothelial cell intercellular junctions, where it functions to facilitate diapedesis, maintain vascular integrity, and transmit survival signals into the cell. Given the importance of PECAM-1-mediated homophilic interactions in mediating each of these cell physiological events, and to reveal the nature and orientation of the PECAM-1-PECAM-1 homophilic-binding interface, we undertook studies aimed at determining the crystal structure of the PECAM-1 homophilic-binding domain, which is composed of amino-terminal immunoglobulin homology domains 1 and 2 (IgD1 and IgD2). The crystal structure revealed that both IgD1 and IgD2 exhibit a classical IgSF fold, having a β-sandwich topology formed by 2 sheets of antiparallel β strands stabilized by the hallmark disulfide bond between the B and F strands. Interestingly, despite previous assignment to the C2 class of immunoglobulin-like domains, the structure of IgD1 reveals that it actually belongs to the I2 set of IgSF folds. Both IgD1 and IgD2 participate importantly in the formation of the trans homophilic-binding interface, with a total buried interface area of >2300 Å(2). These and other unique structural features of PECAM-1 allow for the development of an atomic-level model of the interactions that PECAM-1 forms during assembly of endothelial cell intercellular junctions.
- Blood Research Institute, BloodCenter of Wisconsin, Milwaukee, WI; and.
Organizational Affiliation: