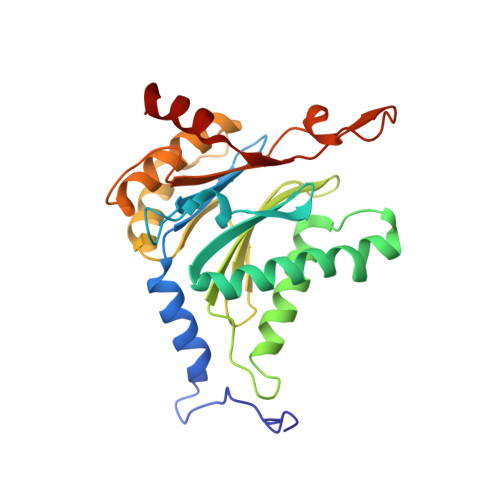
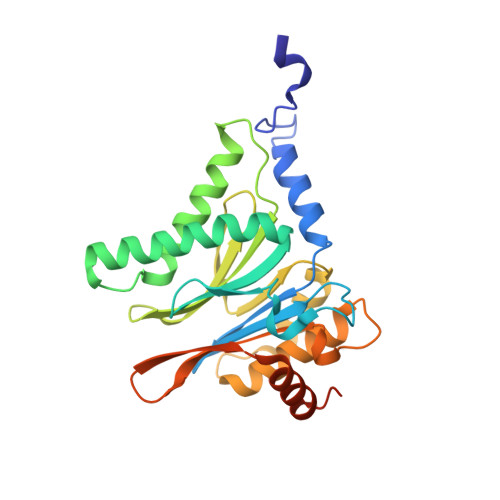
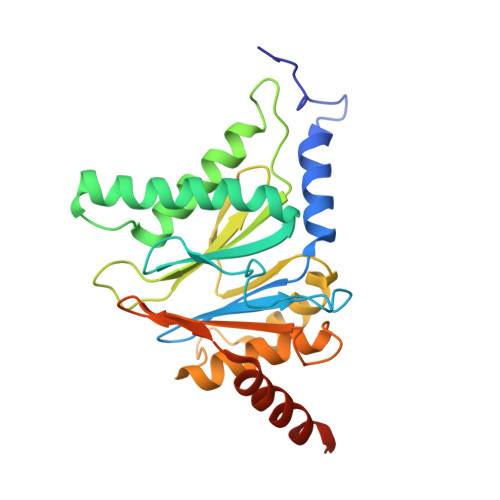
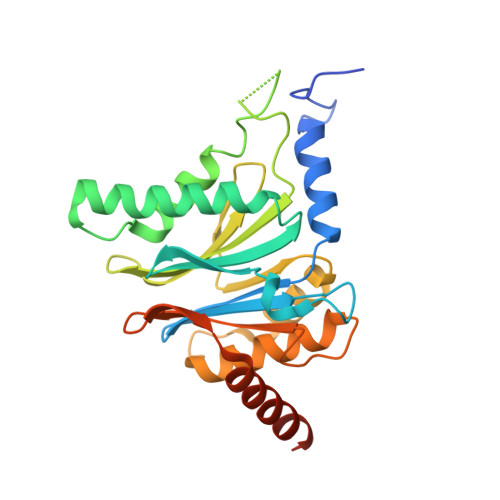
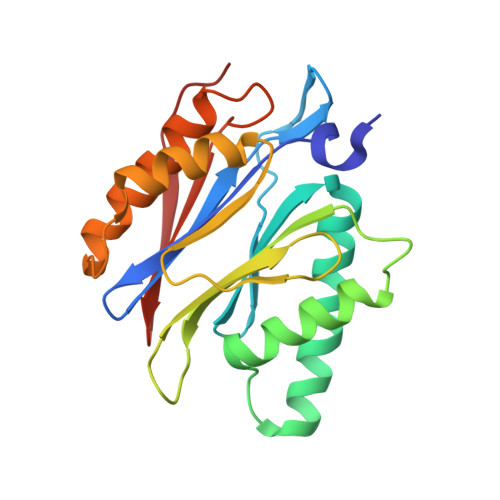
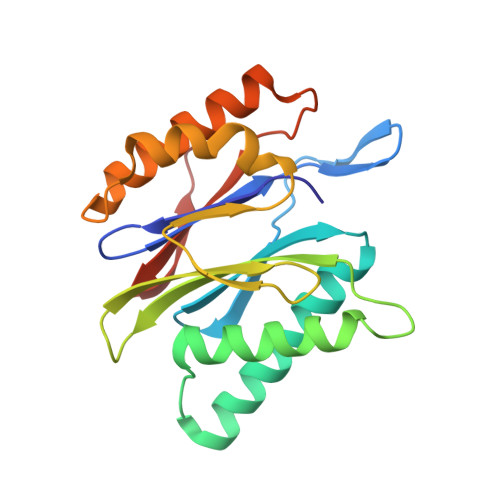
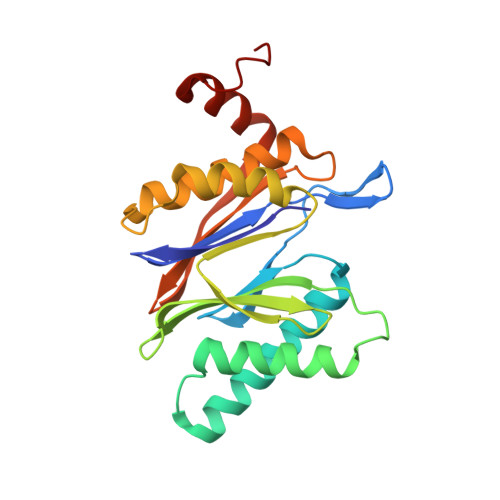
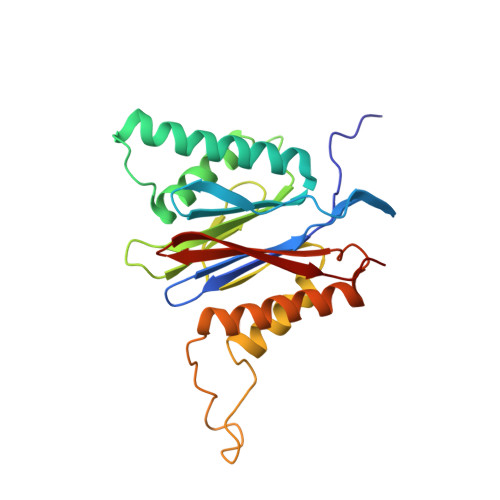
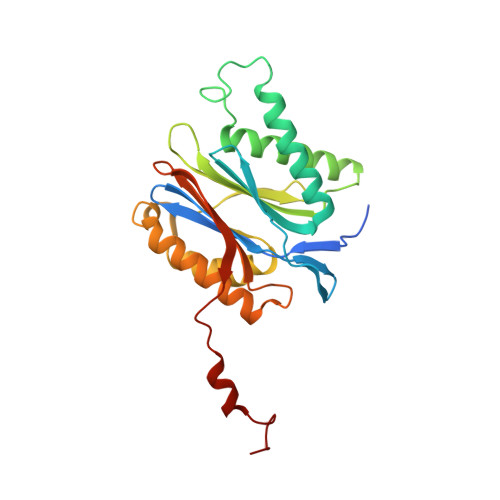
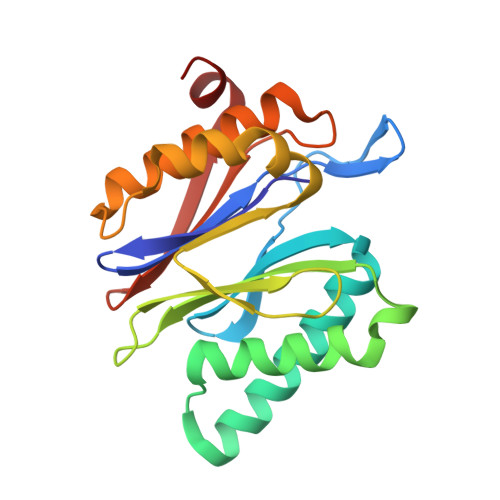
Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Treise, I., Huber, E.M., Klein-Rodewald, T., Heinemeyer, W., Grassmann, S.A., Basler, M., Adler, T., Rathkolb, B., Helming, L., Andres, C., Klaften, M., Landbrecht, C., Wieland, T., Strom, T.M., McCoy, K.D., Macpherson, A.J., Wolf, E., Groettrup, M., Ollert, M., Neff, F., Gailus-Durner, V., Fuchs, H., Hrabe de Angelis, M., Groll, M., Busch, D.H.(2018) Sci Rep 8: 5975-5975
- PubMed: 29654304
- DOI: https://doi.org/10.1038/s41598-018-24199-0
- Primary Citation of Related Structures:
5BXL, 5BXN - PubMed Abstract:
By N-ethyl-N-nitrosourea (ENU) mutagenesis, we generated the mutant mouse line TUB6 that is characterised by severe combined immunodeficiency (SCID) and systemic sterile autoinflammation in homozygotes, and a selective T cell defect in heterozygotes. The causative missense point mutation results in the single amino acid exchange G170W in multicatalytic endopeptidase complex subunit-1 (MECL-1), the β2i-subunit of the immuno- and thymoproteasome. Yeast mutagenesis and crystallographic data suggest that the severe TUB6-phenotype compared to the MECL-1 knockout mouse is caused by structural changes in the C-terminal appendage of β2i that prevent the biogenesis of immuno- and thymoproteasomes. Proteasomes are essential for cell survival, and defective proteasome assembly causes selective death of cells expressing the mutant MECL-1, leading to the severe immunological phenotype. In contrast to the immunosubunits β1i (LMP2) and β5i (LMP7), mutations in the gene encoding MECL-1 have not yet been assigned to human disorders. The TUB6 mutant mouse line exemplifies the involvement of MECL-1 in immunopathogenesis and provides the first mouse model for primary immuno- and thymoproteasome-associated immunodeficiency that may also be relevant in humans.
- Institute for Medical Microbiology, Immunology and Hygiene, Technical University of Munich, Trogerstr. 30, 81675, Munich, Germany.
Organizational Affiliation: