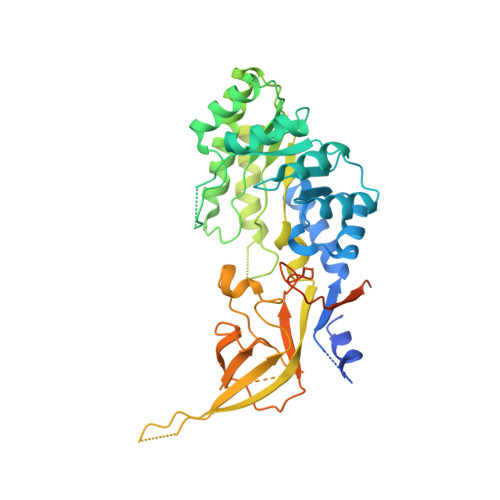
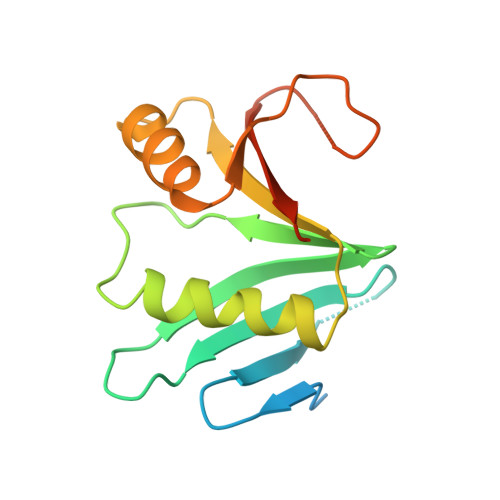
Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1
Wu, D.H., Kaan, H.Y., Zheng, X., Tang, X., He, Y., Vanessa Tan, Q., Zhang, N., Song, H.(2015) Sci Rep 5: 14738-14738
- PubMed: 26443277
- DOI: https://doi.org/10.1038/srep14738
- Primary Citation of Related Structures:
5BWA - PubMed Abstract:
Ornithine decarboxylase (ODC) catalyzes the first and rate-limiting step of polyamine biosynthesis in humans. Polyamines are essential for cell proliferation and are implicated in cellular processes, ranging from DNA replication to apoptosis. Excessive accumulation of polyamines has a cytotoxic effect on cells and elevated level of ODC activity is associated with cancer development. To maintain normal cellular proliferation, regulation of polyamine synthesis is imposed by Antizyme1 (AZ1). The expression of AZ1 is induced by a ribosomal frameshifting mechanism in response to increased intracellular polyamines. AZ1 regulates polyamine homeostasis by inactivating ODC activity and enhancing its degradation. Here, we report the structure of human ODC in complex with N-terminally truncated AZ1 (cAZ1). The structure shows cAZ1 binding to ODC, which occludes the binding of a second molecule of ODC to form the active homodimer. Consequently, the substrate binding site is disrupted and ODC is inactivated. Structural comparison shows that the binding of cAZ1 to ODC causes a global conformational change of ODC and renders its C-terminal region flexible, therefore exposing this region for degradation by the 26S proteasome. Our structure provides the molecular basis for the inactivation of ODC by AZ1 and sheds light on how AZ1 promotes its degradation.
- Institute of Molecular and Cell Biology, 61 Biopolis Drive, Singapore 138673.
Organizational Affiliation: