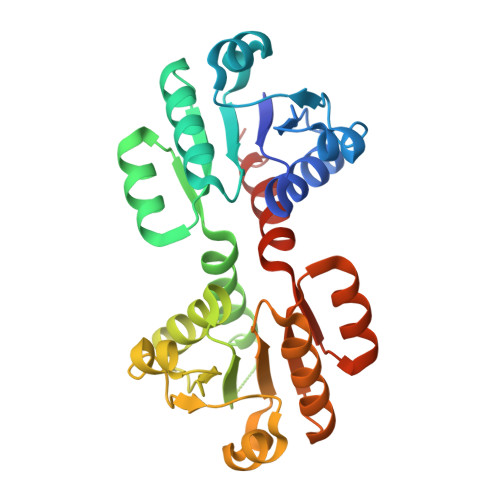
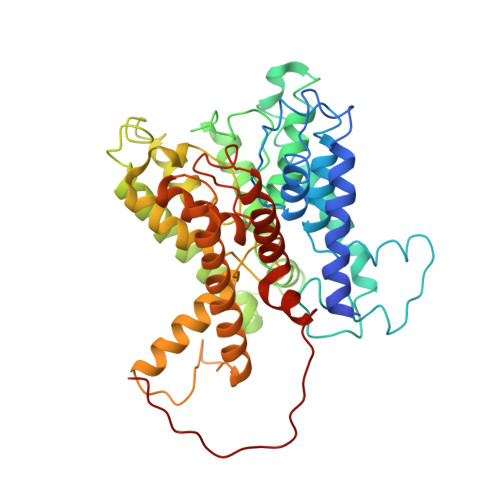
Dissecting the Molecular Mechanism of Nucleotide-Dependent Activation of the KtrAB K+ Transporter.
Szollosi, A., Vieira-Pires, R.S., Teixeira-Duarte, C.M., Rocha, R., Morais-Cabral, J.H.(2016) PLoS Biol 14: e1002356-e1002356
- PubMed: 26771197
- DOI: https://doi.org/10.1371/journal.pbio.1002356
- Primary Citation of Related Structures:
5BUT - PubMed Abstract:
KtrAB belongs to the Trk/Ktr/HKT superfamily of monovalent cation (K+ and Na+) transport proteins that closely resemble K+ channels. These proteins underlie a plethora of cellular functions that are crucial for environmental adaptation in plants, fungi, archaea, and bacteria. The activation mechanism of the Trk/Ktr/HKT proteins remains unknown. It has been shown that ATP stimulates the activity of KtrAB while ADP does not. Here, we present X-ray structural information on the KtrAB complex with bound ADP. A comparison with the KtrAB-ATP structure reveals conformational changes in the ring and in the membrane protein. In combination with a biochemical and functional analysis, we uncover how ligand-dependent changes in the KtrA ring are propagated to the KtrB membrane protein and conclude that, despite their structural similarity, the activation mechanism of KtrAB is markedly different from the activation mechanism of K+ channels.
- IBMC, Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal.
Organizational Affiliation: