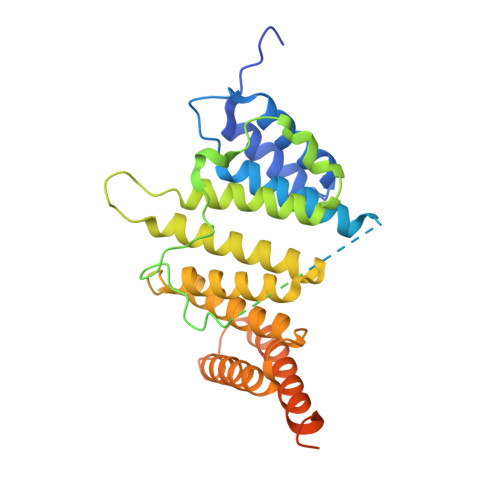
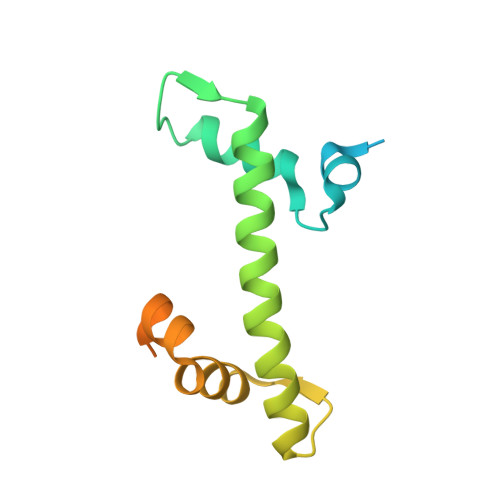
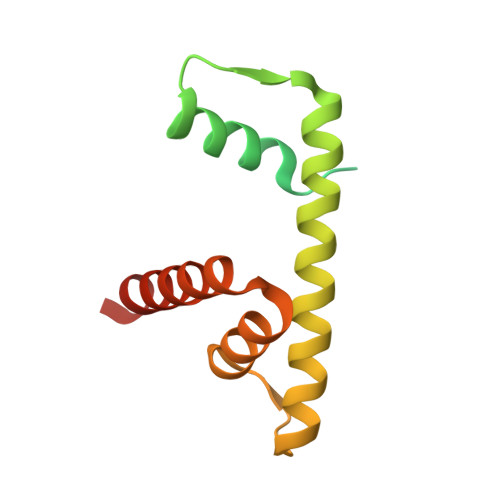
Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Zhang, M., Liu, H., Gao, Y., Zhu, Z., Chen, Z., Zheng, P., Xue, L., Li, J., Teng, M., Niu, L.(2016) Structure 24: 1810-1820
- PubMed: 27618665
- DOI: https://doi.org/10.1016/j.str.2016.08.001
- Primary Citation of Related Structures:
5BT1 - PubMed Abstract:
Histone chaperones are critical for guiding specific post-transcriptional modifications of histones, safeguarding the histone deposition (or disassociation) of nucleosome (dis)assembly, and regulating chromatin structures to change gene activities. HAT1-interacting factor 1 (Hif1) has been reported to be an H3-H4 chaperone and to be involved in telomeric silencing and nucleosome (dis)assembly. However, the structural basis for the interaction of Hif1 with histones remains unknown. Here, we report the complex structure of Hif1 binding to H2A-H2B for uncovering the chaperone specificities of Hif1 on binding to both the H2A-H2B dimer and the H3-H4 tetramer. Our findings reveal that Hif1 interacts with the H2A-H2B dimer and the H3-H4 tetramer via distinct mechanisms, suggesting that Hif1 is a pivotal scaffold on alternate binding of H2A-H2B and H3-H4. These specificities are conserved features of the Sim3-Hif1-NASP interrupted tetratricopeptide repeat proteins, which provide clues for investigating their potential roles in nucleosome (dis)assembly.
- Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China; Key Laboratory of Structural Biology, Chinese Academy of Sciences, Hefei, Anhui 230026, People's Republic of China.
Organizational Affiliation: