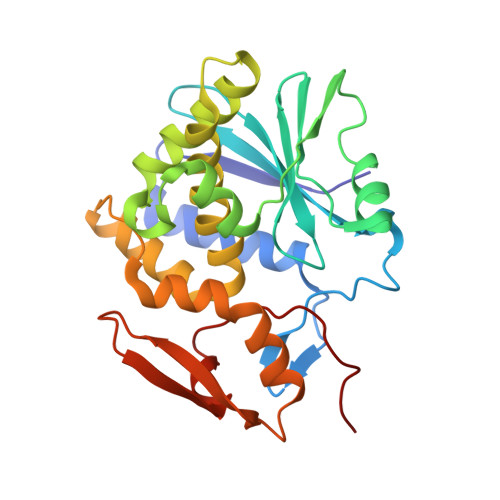
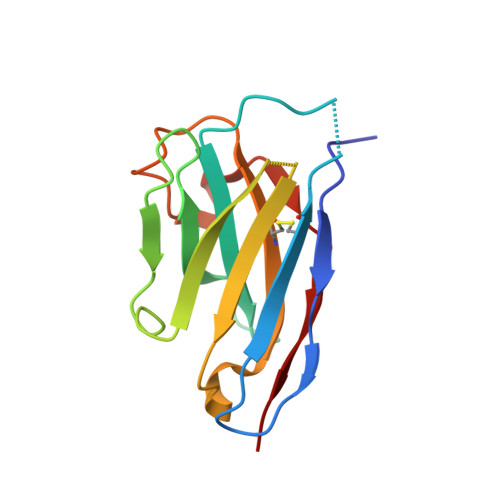
Structural Analysis of Single Domain Antibodies Bound to a Second Neutralizing Hot Spot on Ricin Toxin's Enzymatic Subunit.
Rudolph, M.J., Vance, D.J., Cassidy, M.S., Rong, Y., Mantis, N.J.(2017) J Biological Chem 292: 872-883
- PubMed: 27903650
- DOI: https://doi.org/10.1074/jbc.M116.758102
- Primary Citation of Related Structures:
5BOZ, 5J56, 5J57 - PubMed Abstract:
Ricin toxin is a heterodimer consisting of RTA, a ribosome-inactivating protein, and RTB, a lectin that facilitates receptor-mediated uptake into mammalian cells. In previous studies, we demonstrated that toxin-neutralizing antibodies target four spatially distinct hot spots on RTA, which we refer to as epitope clusters I-IV. In this report, we identified and characterized three single domain camelid antibodies (V H H) against cluster II. One of these V H Hs, V5E1, ranks as one of the most potent ricin-neutralizing antibodies described to date. We solved the X-ray crystal structures of each of the three V H Hs (E1, V1C7, and V5E1) in complex with RTA. V5E1 buries a total of 1,133 Å 2 of surface area on RTA and makes primary contacts with α-helix A (residues 18-32), α-helix F (182-194), as well as the F-G loop. V5E1, by virtue of complementarity determining region 3 (CDR3), may also engage with RTB and potentially interfere with the high affinity galactose-recognition element that plays a critical role in toxin attachment to cell surfaces and intracellular trafficking. The two other V H Hs, E1 and V1C7, bind epitopes adjacent to V5E1 but display only weak toxin neutralizing activity, thereby providing structural insights into specific residues within cluster II that may be critical contact points for toxin inactivation.
- From the New York Structural Biology Center, New York, New York 10027, mrudolph@nysbc.org.
Organizational Affiliation: