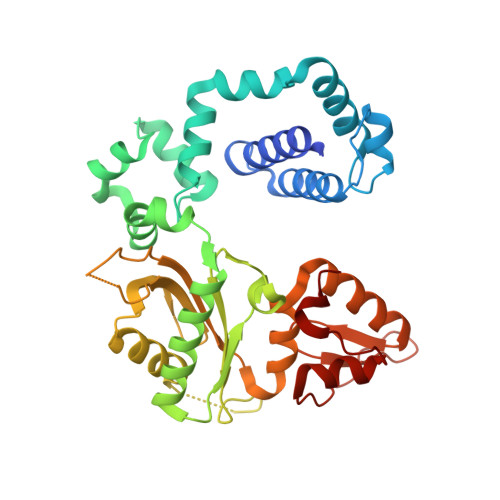
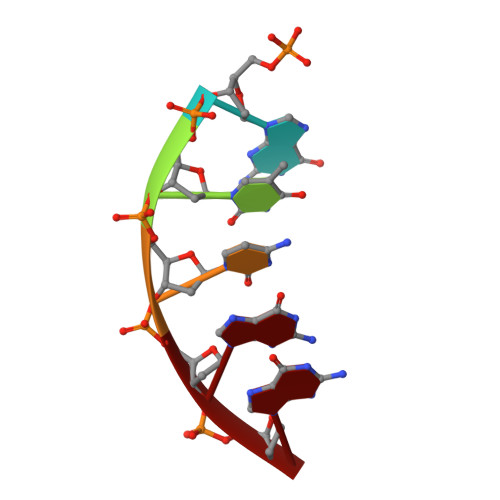
Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Fedeles, B.I., Freudenthal, B.D., Yau, E., Singh, V., Chang, S.C., Li, D., Delaney, J.C., Wilson, S.H., Essigmann, J.M.(2015) Proc Natl Acad Sci U S A 112: E4571-E4580
- PubMed: 26243878
- DOI: https://doi.org/10.1073/pnas.1507709112
- Primary Citation of Related Structures:
5BOL, 5BOM, 5BPC - PubMed Abstract:
During chronic inflammation, neutrophil-secreted hypochlorous acid can damage nearby cells inducing the genomic accumulation of 5-chlorocytosine (5ClC), a known inflammation biomarker. Although 5ClC has been shown to promote epigenetic changes, it has been unknown heretofore if 5ClC directly perpetrates a mutagenic outcome within the cell. The present work shows that 5ClC is intrinsically mutagenic, both in vitro and, at a level of a single molecule per cell, in vivo. Using biochemical and genetic approaches, we have quantified the mutagenic and toxic properties of 5ClC, showing that this lesion caused C→T transitions at frequencies ranging from 3-9% depending on the polymerase traversing the lesion. X-ray crystallographic studies provided a molecular basis for the mutagenicity of 5ClC; a snapshot of human polymerase β replicating across a primed 5ClC-containing template uncovered 5ClC engaged in a nascent base pair with an incoming dATP analog. Accommodation of the chlorine substituent in the template major groove enabled a unique interaction between 5ClC and the incoming dATP, which would facilitate mutagenic lesion bypass. The type of mutation induced by 5ClC, the C→T transition, has been previously shown to occur in substantial amounts both in tissues under inflammatory stress and in the genomes of many inflammation-associated cancers. In fact, many sequence-specific mutational signatures uncovered in sequenced cancer genomes feature C→T mutations. Therefore, the mutagenic ability of 5ClC documented in the present study may constitute a direct functional link between chronic inflammation and the genetic changes that enable and promote malignant transformation.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139; Department Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139; Center for Environmental Health Sciences, Massachusetts Institute of Technology, Cambridge, MA 02139;
Organizational Affiliation: