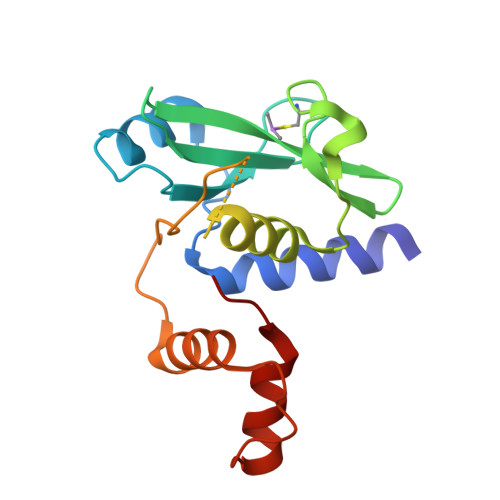
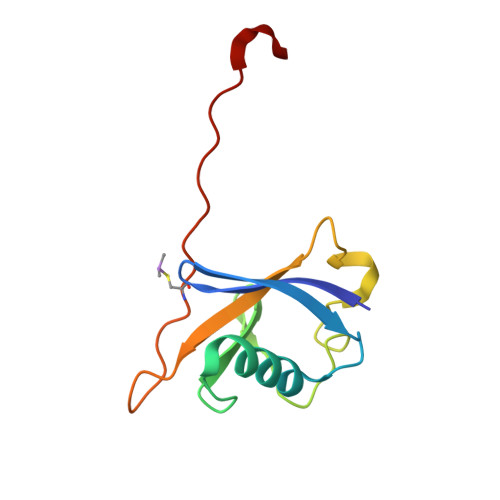
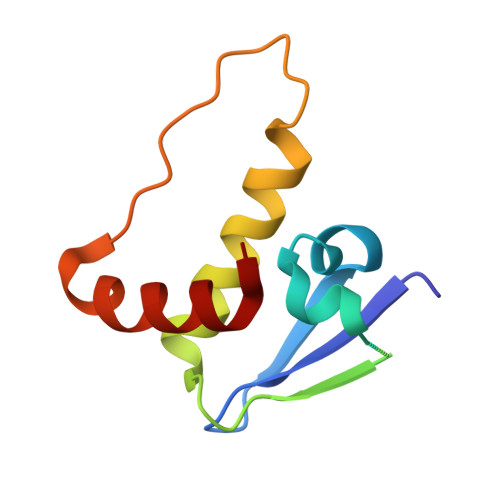
Serendipitous SAD Solution for DMSO-Soaked SOCS2-ElonginC-ElonginB Crystals Using Covalently Incorporated Dimethylarsenic: Insights into Substrate Receptor Conformational Flexibility in Cullin RING Ligases.
Gadd, M.S., Bulatov, E., Ciulli, A.(2015) PLoS One 10: e0131218-e0131218
- PubMed: 26121586
- DOI: https://doi.org/10.1371/journal.pone.0131218
- Primary Citation of Related Structures:
5BO4 - PubMed Abstract:
Suppressor of cytokine signalling 2 (SOCS2) is the substrate-binding component of a Cullin-RING E3 ubiquitin ligase (CRL) complex that targets phosphorylated hormone receptors for degradation by the ubiquitin-proteasome system. As a key regulator of the transcriptional response to growth signals, SOCS2 and its protein complex partners are potential targets for small molecule development. We found that crystals of SOCS2 in complex with its adaptor proteins, Elongin C and Elongin B, underwent a change in crystallographic parameters when treated with dimethyl sulfoxide during soaking experiments. To solve the phase problem for the new crystal form we identified the presence of arsenic atoms in the crystals, a result of covalent modification of cysteines by cacodylate, and successfully extracted anomalous signal from these atoms for experimental phasing. The resulting structure provides a means for solving future structures where the crystals must be treated with DMSO for ligand soaking approaches. Additionally, the conformational changes induced in this structure reveal flexibility within SOCS2 that match those postulated by previous molecular dynamics simulations. This conformational flexibility illustrates how SOCS2 can orient its substrates for successful ubiquitination by other elements of the CRL complex.
- Division of Biological Chemistry and Drug Discovery, College of Life Sciences, University of Dundee, James Black Centre, Dow Street, Dundee, United Kingdom.
Organizational Affiliation: