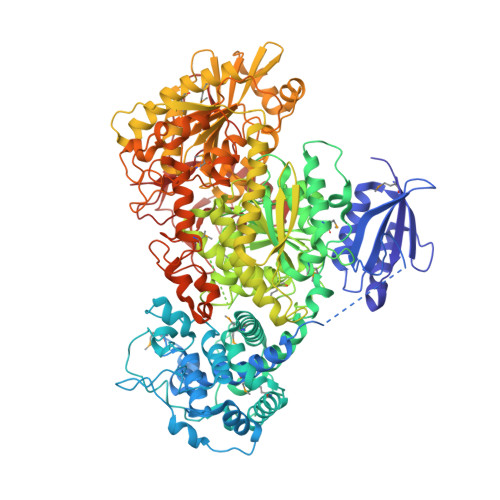
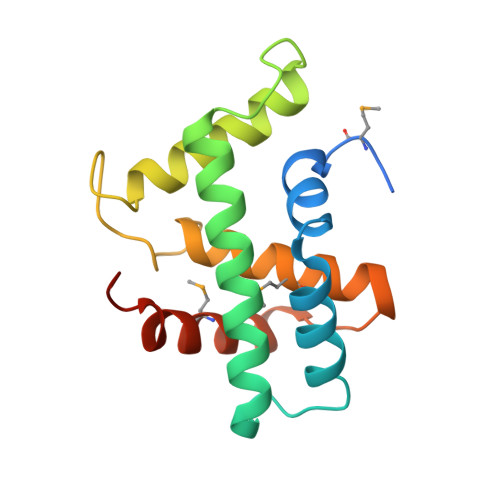
Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3
Wang, X., Yao, D., Xu, J.G., Li, A.R., Xu, J., Fu, P., Zhou, Y., Zhu, Y.(2016) Nat Struct Mol Biol 23: 868-870
- PubMed: 27455460
- DOI: https://doi.org/10.1038/nsmb.3269
- Primary Citation of Related Structures:
5B7I - PubMed Abstract:
Bacteriophages express proteins that inactivate the CRISPR-Cas bacterial immune system. Here we report the crystal structure of the anti-CRISPR protein AcrF3 in complex with Pseudomonas aeruginosa Cas3 (PaCas3). AcrF3 forms a homodimer that locks PaCas3 in an ADP-bound form, blocks the entrance of the DNA-binding tunnel in the helicase domain, and masks the linker region and C-terminal domain of PaCas3, thereby preventing recruitment by Cascade and inhibiting the type I-F CRISPR-Cas system.
- Life Sciences Institute and Innovation Center for Cell Signaling Network, Zhejiang University, Hangzhou, China.
Organizational Affiliation: