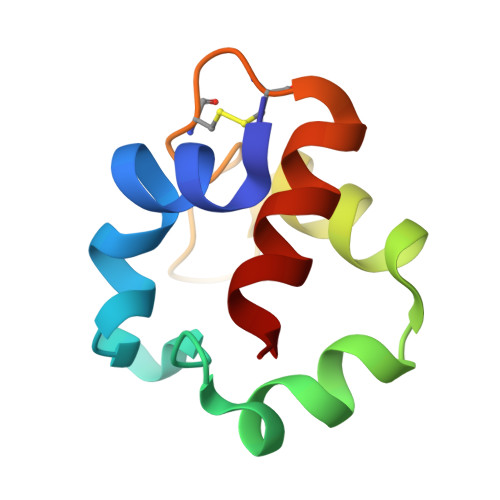
Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Masanari, M., Fujii, S., Kawahara, K., Oki, H., Tsujino, H., Maruno, T., Kobayashi, Y., Ohkubo, T., Wakai, S., Sambongi, Y.(2016) Biosci Biotechnol Biochem
- PubMed: 27648635
- DOI: https://doi.org/10.1080/09168451.2016.1232155
- Primary Citation of Related Structures:
5B6Q - PubMed Abstract:
Monomeric cytochrome c 5 from deep-sea piezophilic Shewanella violacea (SVcytc 5 ) was stable against heat and denaturant compared with the homologous protein from shallow-sea piezo-sensitive Shewanella livingstonensis (SLcytc 5 ). Here, the SVcytc 5 crystal structure revealed that the Lys-50 side chain on the flexible loop formed a hydrogen bond with heme whereas that of corresponding hydrophobic Leu-50 could not form such a bond in SLcytc 5 , which appeared to be one of possible factors responsible for the difference in stability between the two proteins. This structural insight was confirmed by a reciprocal mutagenesis study on the thermal stability of these two proteins. As SVcytc 5 was isolated from a deep-sea piezophilic bacterium, the present comparative study indicates that adaptation of monomeric SVcytc 5 to high pressure environments results in stabilization against heat.
- a Graduate School of Biosphere Science , Hiroshima University , Higashi-Hiroshima , Japan.
Organizational Affiliation: