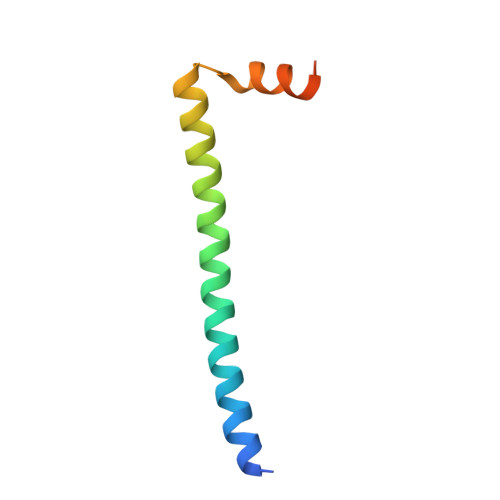
Structural similarities and differences in H-NS family proteins revealed by the N-terminal structure of TurB in Pseudomonas putida KT2440
Suzuki-Minakuchi, C., Kawazuma, K., Matsuzawa, J., Vasileva, D., Fujimoto, Z., Terada, T., Okada, K., Nojiri, H.(2016) FEBS Lett 590: 3583-3594
- PubMed: 27709616
- DOI: https://doi.org/10.1002/1873-3468.12425
- Primary Citation of Related Structures:
5B52 - PubMed Abstract:
H-NS family proteins play key roles in bacterial nucleoid compaction and global transcription. MvaT homologues in Pseudomonas have almost negligible amino acid sequence identity with H-NS, but can complement an hns-related phenotype of Escherichia coli. Here, we report the crystal structure of the N-terminal dimerization/oligomerization domain of TurB, an MvaT homologue in Pseudomonas putida KT2440. Our data identify two dimerization sites; the structure of the central dimerization site is almost the same as the corresponding region of H-NS, whereas the terminal dimerization sites are different. Our results reveal similarities and differences in dimerization and oligomerization mechanisms between H-NS and TurB.
- Biotechnology Research Center, The University of Tokyo, Japan. ucsmina@mail.ecc.u-tokyo.ac.jp.
Organizational Affiliation: