Beta-sheet elasticity of peptide self-assembly mimic, PSAM, with a grafted sequence characterized by comprehensive analyses of isomorphous crystals
Fujiwara, H., Hongo, K., Hori, Y., Yoshida, N., Makabe, K.(2019) J Mol Liq 290
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2019) J Mol Liq 290
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer surface protein A | A [auth O] | 251 | Borreliella burgdorferi | Mutation(s): 18 Gene Names: ospA | 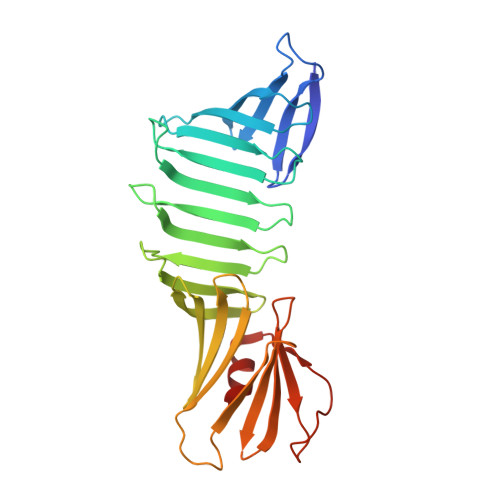 |
UniProt | |||||
Find proteins for P0CL66 (Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31)) Explore P0CL66 Go to UniProtKB: P0CL66 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CL66 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 33.37 | α = 90 |
| b = 53.658 | β = 100.35 |
| c = 65.756 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Coot | model building |