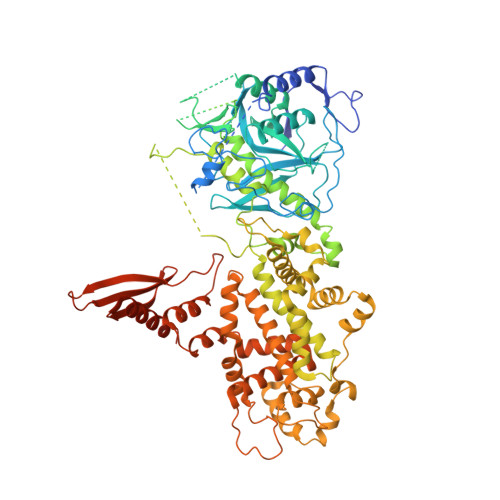
Structure of Human DROSHA
Kwon, S.C., Nguyen, T.A., Choi, Y.G., Jo, M.H., Hohng, S., Kim, V.N., Woo, J.S.(2016) Cell 164: 81-90
- PubMed: 26748718
- DOI: https://doi.org/10.1016/j.cell.2015.12.019
- Primary Citation of Related Structures:
5B16 - PubMed Abstract:
MicroRNA maturation is initiated by RNase III DROSHA that cleaves the stem loop of primary microRNA. DROSHA functions together with its cofactor DGCR8 in a heterotrimeric complex known as Microprocessor. Here, we report the X-ray structure of DROSHA in complex with the C-terminal helix of DGCR8. We find that DROSHA contains two DGCR8-binding sites, one on each RNase III domain (RIIID), which mediate the assembly of Microprocessor. The overall structure of DROSHA is surprisingly similar to that of Dicer despite no sequence homology apart from the C-terminal part, suggesting that DROSHA may have evolved from a Dicer homolog. DROSHA exhibits unique features, including non-canonical zinc-finger motifs, a long insertion in the first RIIID, and the kinked link between Connector helix and RIIID, which explains the 11-bp-measuring "ruler" activity of DROSHA. Our study implicates the evolutionary origin of DROSHA and elucidates the molecular basis of Microprocessor assembly and primary microRNA processing.
- Center for RNA Research, Institute for Basic Science, Seoul 08826, Korea; School of Biological Sciences, Seoul National University, Seoul 08826, Korea.
Organizational Affiliation: