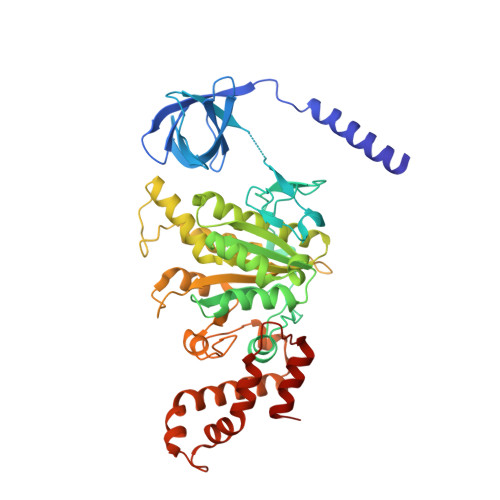
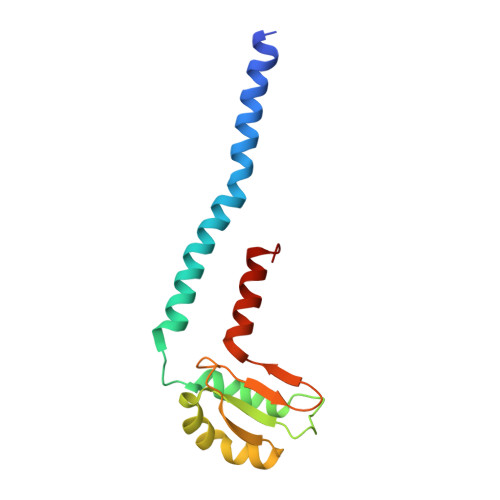
Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Imada, K., Minamino, T., Uchida, Y., Kinoshita, M., Namba, K.(2016) Proc Natl Acad Sci U S A 113: 3633-3638
- PubMed: 26984495
- DOI: https://doi.org/10.1073/pnas.1524025113
- Primary Citation of Related Structures:
5B0O - PubMed Abstract:
FliI and FliJ form the FliI6FliJ ATPase complex of the bacterial flagellar export apparatus, a member of the type III secretion system. The FliI6FliJ complex is structurally similar to the α3β3γ complex of F1-ATPase. The FliH homodimer binds to FliI to connect the ATPase complex to the flagellar base, but the details are unknown. Here we report the structure of the homodimer of a C-terminal fragment of FliH (FliHC2) in complex with FliI. FliHC2 shows an unusually asymmetric homodimeric structure that markedly resembles the peripheral stalk of the A/V-type ATPases. The FliHC2-FliI hexamer model reveals that the C-terminal domains of the FliI ATPase face the cell membrane in a way similar to the F/A/V-type ATPases. We discuss the mechanism of flagellar ATPase complex formation and a common origin shared by the type III secretion system and the F/A/V-type ATPases.
- Department of Macromolecular Science, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, Japan; kimada@chem.sci.osaka-u.ac.jp.
Organizational Affiliation: