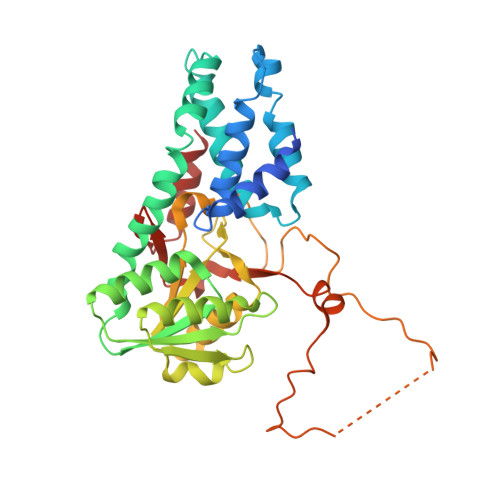
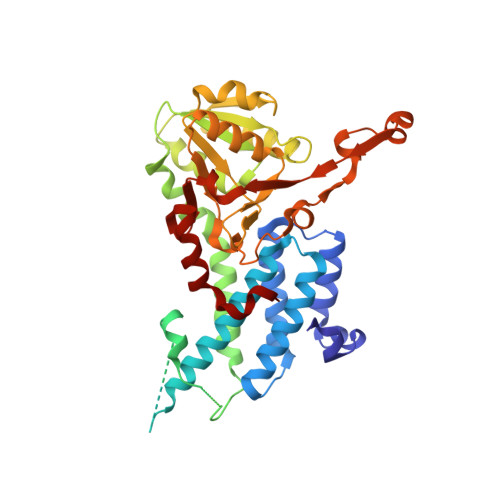
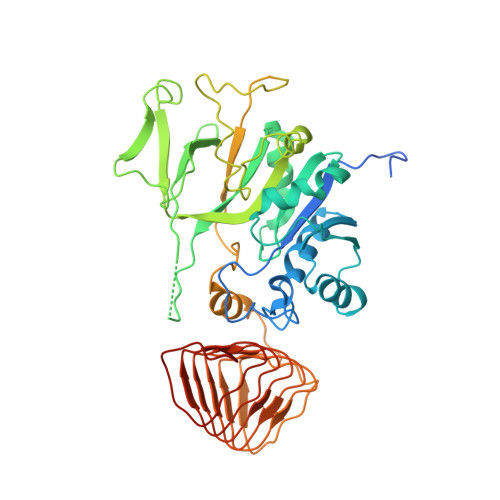
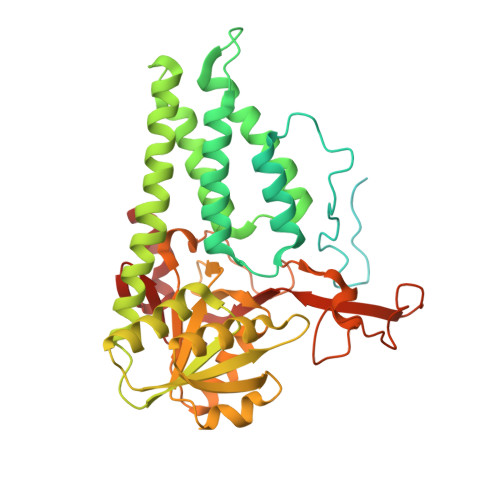
Crystal structure of eukaryotic translation initiation factor 2B
Kashiwagi, K., Takahashi, M., Nishimoto, M., Hiyama, T.B., Higo, T., Umehara, T., Sakamoto, K., Ito, T., Yokoyama, S.(2016) Nature 531: 47-52
- PubMed: 26901872
- DOI: https://doi.org/10.1038/nature16991
- Primary Citation of Related Structures:
5B04 - PubMed Abstract:
Eukaryotic cells restrict protein synthesis under various stress conditions, by inhibiting the eukaryotic translation initiation factor 2B (eIF2B). eIF2B is the guanine nucleotide exchange factor for eIF2, a heterotrimeric G protein consisting of α-, β- and γ-subunits. eIF2B exchanges GDP for GTP on the γ-subunit of eIF2 (eIF2γ), and is inhibited by stress-induced phosphorylation of eIF2α. eIF2B is a heterodecameric complex of two copies each of the α-, β-, γ-, δ- and ε-subunits; its α-, β- and δ-subunits constitute the regulatory subcomplex, while the γ- and ε-subunits form the catalytic subcomplex. The three-dimensional structure of the entire eIF2B complex has not been determined. Here we present the crystal structure of Schizosaccharomyces pombe eIF2B with an unprecedented subunit arrangement, in which the α2β2δ2 hexameric regulatory subcomplex binds two γε dimeric catalytic subcomplexes on its opposite sides. A structure-based in vitro analysis by a surface-scanning site-directed photo-cross-linking method identified the eIF2α-binding and eIF2γ-binding interfaces, located far apart on the regulatory and catalytic subcomplexes, respectively. The eIF2γ-binding interface is located close to the conserved 'NF motif', which is important for nucleotide exchange. A structural model was constructed for the complex of eIF2B with phosphorylated eIF2α, which binds to eIF2B more strongly than the unphosphorylated form. These results indicate that the eIF2α phosphorylation generates the 'nonproductive' eIF2-eIF2B complex, which prevents nucleotide exchange on eIF2γ, and thus provide a structural framework for the eIF2B-mediated mechanism of stress-induced translational control.
- Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: