Structural Basis for Heme Recognition by HmuT Responsible for Heme Transport to the Heme Transporter in Corynebacterium glutamicum
Muraki, N., Aono, S.(2015) Chem Lett
Experimental Data Snapshot
Starting Model: experimental
View more details
(2015) Chem Lett
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ABC-type transporter, periplasmic component | 344 | Corynebacterium glutamicum ATCC 13032 | Mutation(s): 0 Gene Names: Cgl0389 | 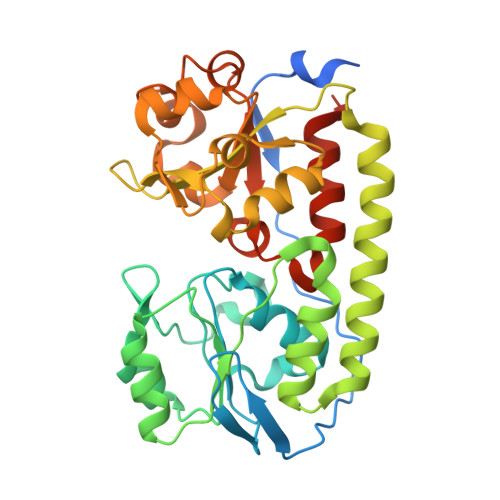 | |
UniProt | |||||
Find proteins for Q8NTB8 (Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / JCM 1318 / BCRC 11384 / CCUG 27702 / LMG 3730 / NBRC 12168 / NCIMB 10025 / NRRL B-2784 / 534)) Explore Q8NTB8 Go to UniProtKB: Q8NTB8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8NTB8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | B [auth A] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.04 | α = 90 |
| b = 73.04 | β = 90 |
| c = 147.32 | γ = 90 |
| Software Name | Purpose |
|---|---|
| iMOSFLM | data processing |
| SCALA | data scaling |
| PHASER | phasing |
| Coot | model building |
| PHENIX | refinement |