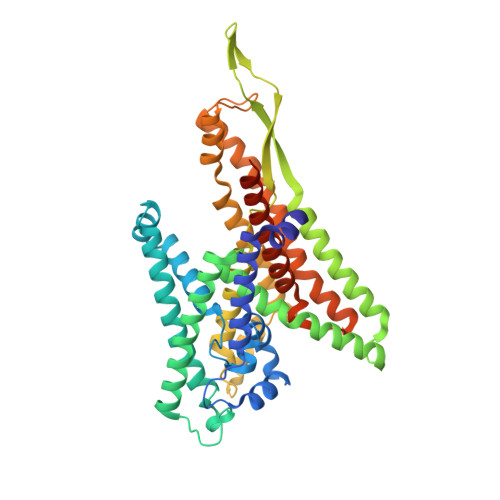
Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Tanaka, Y., Sugano, Y., Takemoto, M., Mori, T., Furukawa, A., Kusakizako, T., Kumazaki, K., Kashima, A., Ishitani, R., Sugita, Y., Nureki, O., Tsukazaki, T.(2015) Cell Rep 13: 1561-1568
- PubMed: 26586438
- DOI: https://doi.org/10.1016/j.celrep.2015.10.025
- Primary Citation of Related Structures:
5AWW, 5CH4 - PubMed Abstract:
The bacterial SecYEG translocon functions as a conserved protein-conducting channel. Conformational transitions of SecYEG allow protein translocation across the membrane without perturbation of membrane permeability. Here, we report the crystal structures of intact SecYEG at 2.7-Å resolution and of peptide-bound SecYEG at 3.6-Å resolution. The higher-resolution structure revealed that the cytoplasmic loop of SecG covers the hourglass-shaped channel, which was confirmed to also occur in the membrane by disulfide bond formation analysis and molecular dynamics simulation. The cytoplasmic loop may be involved in protein translocation. In addition, the previously unknown peptide-bound crystal structure of SecYEG implies that interactions between the cytoplasmic side of SecY and signal peptides are related to lateral gate opening at the first step of protein translocation. These SecYEG structures therefore provide a number of structural insights into the Sec machinery for further study.
- Department of Systems Biology, Graduate School of Biological Sciences, Nara Institute of Science and Technology, 8916-5, Takayama-cho, Ikoma, Nara 630-0192, Japan.
Organizational Affiliation: