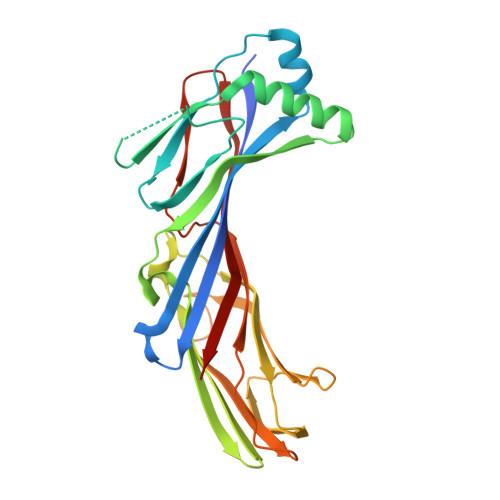
Structural basis for the recognition of two consecutive mutually interacting DPF motifs by the SGIP1 mu homology domain.
Shimada, A., Yamaguchi, A., Kohda, D.(2016) Sci Rep 6: 19565-19565
- PubMed: 26822536
- DOI: https://doi.org/10.1038/srep19565
- Primary Citation of Related Structures:
5AWR, 5AWS, 5AWT, 5AWU - PubMed Abstract:
FCHo1, FCHo2, and SGIP1 are key regulators of clathrin-mediated endocytosis. Their μ homology domains (μHDs) interact with the C-terminal region of an endocytic scaffold protein, Eps15, containing fifteen Asp-Pro-Phe (DPF) motifs. Here, we show that the high-affinity μHD-binding site in Eps15 is a region encompassing six consecutive DPF motifs, while the minimal μHD-binding unit is two consecutive DPF motifs. We present the crystal structures of the SGIP1 μHD in complex with peptides containing two DPF motifs. The peptides bind to a novel ligand-binding site of the μHD, which is distinct from those of other distantly related μHD-containing proteins. The two DPF motifs, which adopt three-dimensional structures stabilized by sequence-specific intramotif and intermotif interactions, are extensively recognized by the μHD and are both required for binding. Thus, consecutive and singly scattered DPF motifs play distinct roles in μHD binding.
- Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Organizational Affiliation: