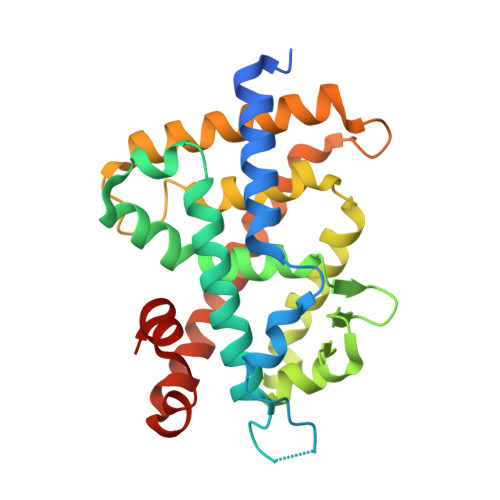
Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Anami, Y., Sakamaki, Y., Itoh, T., Inaba, Y., Nakabayashi, M., Ikura, T., Ito, N., Yamamoto, K.(2015) Bioorg Med Chem 23: 7274-7281
- PubMed: 26515040
- DOI: https://doi.org/10.1016/j.bmc.2015.10.026
- Primary Citation of Related Structures:
5AWJ, 5AWK - PubMed Abstract:
1α,25-Dihydroxyvitamin D3 exerts its actions by binding to vitamin D receptor (VDR). We are continuing the study related to the alteration of pocket structure of VDR by 22-alkyl substituent of ligands and the relationships between the alteration and agonistic/antagonistic activity. Previously we reported that compounds 2 (22-H), 3 (22S-Et), and 4 (22S-Bu) are VDR agonist, partial agonist and antagonist, respectively. Here, we describe the synthesis and biological evaluation of 22S-hexyl analog 5 (22S-Hex), which was designed to be a stronger VDR antagonist than 4. Unexpectedly, 5 showed partial agonistic but not antagonistic activity when bound to VDR, indicating that it is not necessarily true that the bulkier the side chain is, the stronger the antagonistic activity will be. X-ray crystallographic analysis of the VDR-ligand-binding domain (VDR-LBD) accommodating compound 5 indicated that the partial agonist activity of 5 is dependent on the mixed population of the agonistic and antagonistic conformations. Binding of compound 5 may not bring the complex into the only antagonistic conformation due to the large conformational change of the VDR-LBD. From this study it was found that fine tuning of agonistic/antagonistic activity for VDR is possible by 22-alkyl chain length of ligands.
- Laboratory of Drug Design and Medicinal Chemistry, Showa Pharmaceutical University, 3-3165 Higashi-Tamagawagakuen, Machida, Tokyo 194-8543, Japan.
Organizational Affiliation: