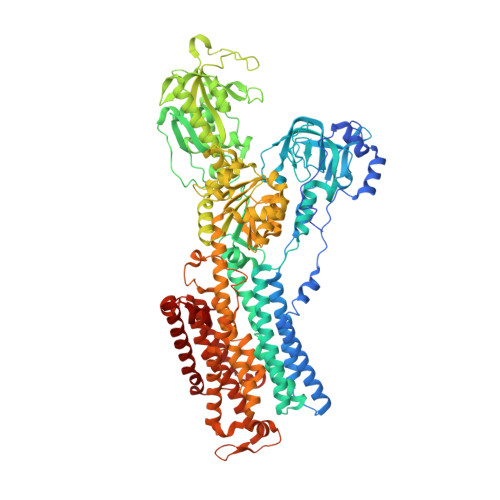
Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Ogawa, H., Cornelius, F., Hirata, A., Toyoshima, C.(2015) Nat Commun 6: 8004-8004
- PubMed: 26258479
- DOI: https://doi.org/10.1038/ncomms9004
- Primary Citation of Related Structures:
5AVQ, 5AVR, 5AVS, 5AVT, 5AVU, 5AVV, 5AVW, 5AVX, 5AVY, 5AVZ, 5AW0, 5AW1, 5AW2, 5AW3, 5AW4, 5AW5, 5AW6, 5AW7, 5AW8, 5AW9 - PubMed Abstract:
Na(+),K(+)-ATPase transfers three Na(+) from the cytoplasm into the extracellular medium and two K(+) in the opposite direction per ATP hydrolysed. The binding and release of Na(+) and K(+) are all thought to occur sequentially. Here we demonstrate by X-ray crystallography of the ATPase in E2·MgF4(2-)·2K(+), a state analogous to E2·Pi·2K(+), combined with isotopic measurements, that the substitution of the two K(+) with congeners in the extracellular medium indeed occurs at different rates, substantially faster at site II. An analysis of thermal movements of protein atoms in the crystal shows that the M3-M4E helix pair opens and closes the ion pathway leading to the extracellular medium, allowing K(+) at site II to be substituted first. Taken together, these results indicate that site I K(+) is the first cation to bind to the empty cation-binding sites after releasing three Na(+).
- Institute of Molecular and Cellular Biosciences, The University of Tokyo, Bunkyo-ku, Tokyo 113-0032, Japan.
Organizational Affiliation: