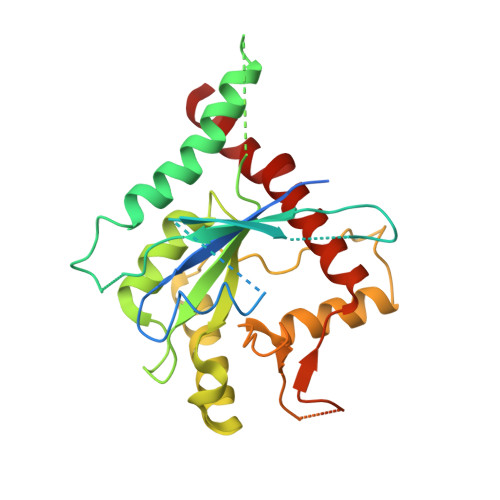
Crystal Structure of Cdc11, a Septin Subunit from Saccharomyces Cerevisiae.
Brausemann, A., Gerhardt, S., Schott, A.K., Einsle, O., Grosse-Berkenbusch, A., Johnsson, N., Gronemeyer, T.(2016) J Struct Biol 193: 157
- PubMed: 26780475
- DOI: https://doi.org/10.1016/j.jsb.2016.01.004
- Primary Citation of Related Structures:
5AR1 - PubMed Abstract:
Septins are a conserved family of GTP-binding proteins that assemble into a highly ordered array of filaments at the mother bud neck in Saccharomyces cerevisiae cells. Many molecular functions and mechanisms of the septins in S. cerevisiae were already uncovered. However, structural information is only available from modeling the crystallized subunits of the human septins into the EM cryomicroscopy data of the yeast hetero-octameric septin rod. Octameric rods are the building block of septin filaments in yeast. We present here the first crystal structure of Cdc11, the terminal subunit of the octameric rod and discuss its structure in relation to its human homologues. Size exclusion chromatography analysis revealed that Cdc11 forms homodimers through its C-terminal coiled coil tail.
- Institute for Biochemistry, Albert-Ludwigs University, 79104 Freiburg, Germany.
Organizational Affiliation: