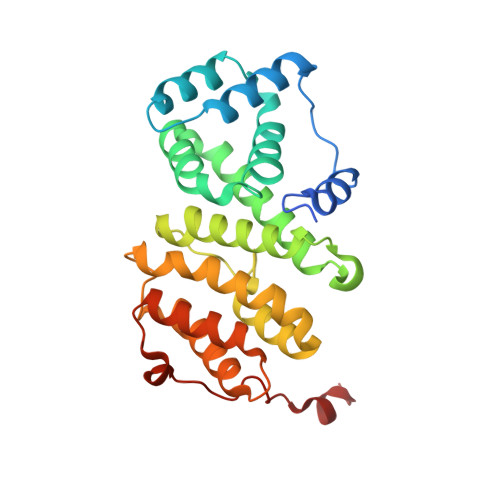
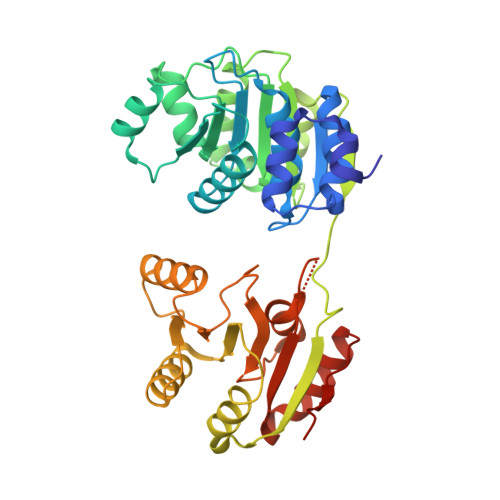
Structure of a Human 4E-T/Ddx6/Cnot1 Complex Reveals the Different Interplay of Ddx6-Binding Proteins with the Ccr4-not Complex.
Ozgur, S., Basquin, J., Kamenska, A., Filipowicz, W., Standart, N., Conti, E.(2015) Cell Rep 13: 703
- PubMed: 26489469
- DOI: https://doi.org/10.1016/j.celrep.2015.09.033
- Primary Citation of Related Structures:
5ANR - PubMed Abstract:
The DEAD-box protein DDX6 is a central component of translational repression mechanisms in maternal mRNA storage in oocytes and microRNA-mediated silencing in somatic cells. DDX6 interacts with the CCR4-NOT complex and functions in concert with several post-transcriptional regulators, including Edc3, Pat1, and 4E-T. We show that the conserved CUP-homology domain (CHD) of human 4E-T interacts directly with DDX6 in both the presence and absence of the central MIF4G domain of CNOT1. The 2.1-Å resolution structure of the corresponding ternary complex reveals how 4E-T CHD wraps around the RecA2 domain of DDX6 and contacts CNOT1. Although 4E-T CHD lacks recognizable sequence similarity with Edc3 or Pat1, it shares the same DDX6-binding surface. In contrast to 4E-T, however, the Edc3 and Pat1 FDF motifs dissociate from DDX6 upon CNOT1 MIF4G binding in vitro. The results underscore the presence of a complex network of simultaneous and/or mutually exclusive interactions in DDX6-mediated repression.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried/Munich, Germany.
Organizational Affiliation: