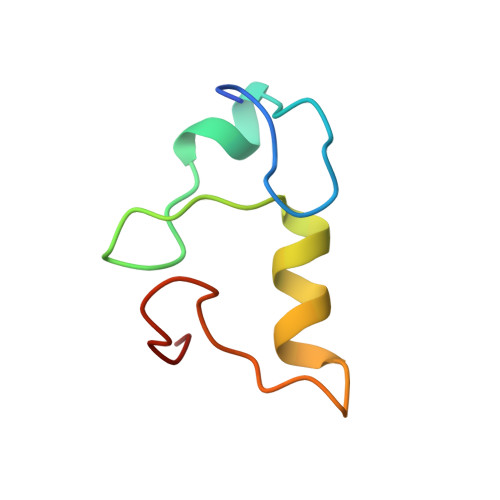
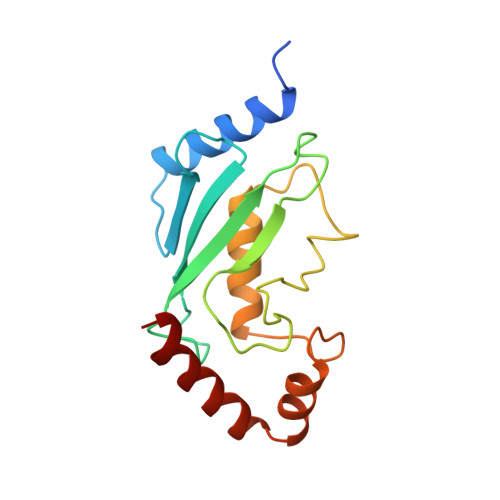
Architecture of the Ubiquitylation Module of the Yeast Ccr4-not Complex.
Bhaskar, V., Basquin, J., Conti, E.(2015) Structure 23: 921
- PubMed: 25914052
- DOI: https://doi.org/10.1016/j.str.2015.03.011
- Primary Citation of Related Structures:
5AIE, 5AJD - PubMed Abstract:
The Ccr4-Not complex regulates eukaryotic gene expression at multiple levels, including mRNA turnover, translational repression, and transcription. We have studied the ubiquitylation module of the yeast Ccr4-Not complex and addressed how E3 ligase binds cognate E2 and how it is tethered to the complex. The 2.8-Å resolution crystal structure of the N-terminal RING domain of Not4 in complex with Ubc4 shows the detailed interactions of this E3-E2 complex. The 3.6-Å resolution crystal structure of the C-terminal domain of the yeast Not4 in complex with the C-terminal domain of Not1 reveals how a largely extended region at the C-terminus of Not4 wraps around a HEAT-repeat region of Not1. This C-terminal region of Not4 is only partly conserved in metazoans, rationalizing its weaker Not1-binding properties. The structural and biochemical data show how Not1 can incorporate both the ubiquitylation module and the Not2-Not3/5 module concomitantly in the Ccr4-Not complex.
- Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, 82152 Munich, Germany.
Organizational Affiliation: