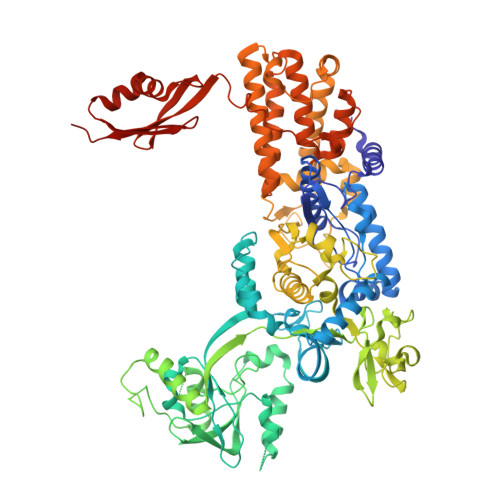
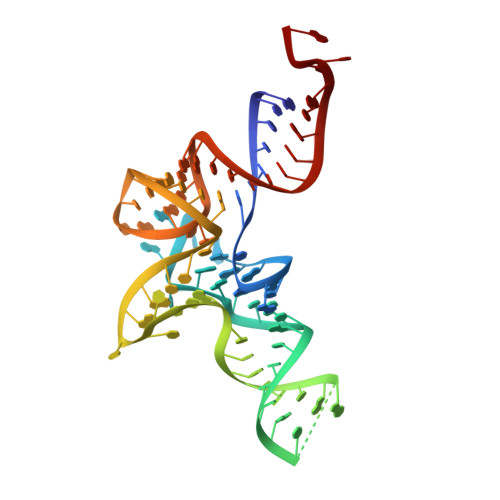Structural Characterization of Antibiotic Self-Immunity tRNA Synthetase in Plant Tumour Biocontrol Agent.
Chopra, S., Palencia, A., Virus, C., Schulwitz, S., Temple, B.R., Cusack, S., Reader, J.(2016) Nat Commun 7: 12928
- PubMed: 27713402
- DOI: https://doi.org/10.1038/ncomms12928
- Primary Citation of Related Structures:
5AH5 - PubMed Abstract:
Antibiotic-producing microbes evolved self-resistance mechanisms to avoid suicide. The biocontrol Agrobacterium radiobacter K84 secretes the Trojan Horse antibiotic agrocin 84 that is selectively transported into the plant pathogen A. tumefaciens and processed into the toxin TM84. We previously showed that TM84 employs a unique tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase (LeuRS), while the TM84-producer prevents self-poisoning by expressing a resistant LeuRS AgnB2. We now identify a mechanism by which the antibiotic-producing microbe resists its own toxin. Using a combination of structural, biochemical and biophysical approaches, we show that AgnB2 evolved structural changes so as to resist the antibiotic by eliminating the tRNA-dependence of TM84 binding. Mutagenesis of key resistance determinants results in mutants adopting an antibiotic-sensitive phenotype. This study illuminates the evolution of resistance in self-immunity genes and provides mechanistic insights into a fascinating tRNA-dependent antibiotic with applications for the development of anti-infectives and the prevention of biocontrol emasculation.
- Department of Cell Biology and Physiology, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599-7090, USA.
Organizational Affiliation: