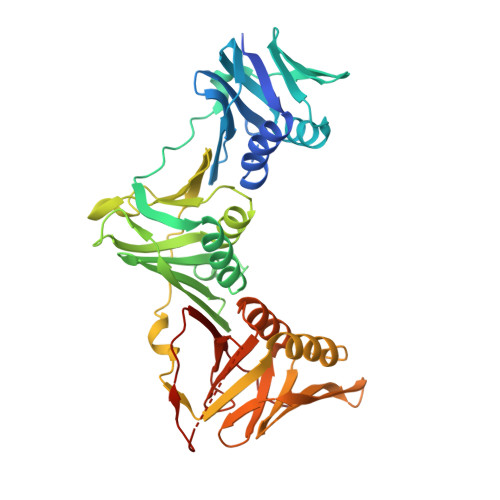
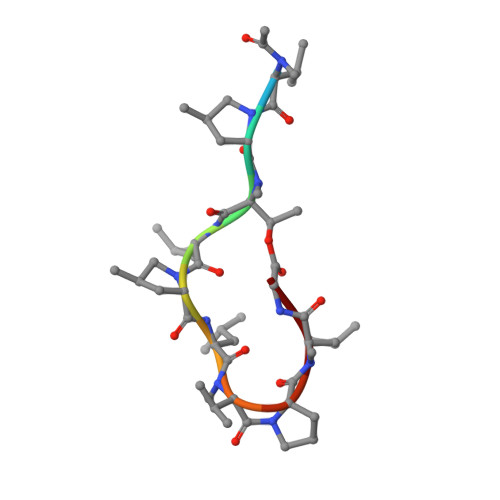
Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Kling, A., Lukat, P., Almeida, D.V., Bauer, A., Fontaine, E., Sordello, S., Zaburannyi, N., Herrmann, J., Wenzel, S.C., Konig, C., Ammerman, N.C., Barrio, M.B., Borchers, K., Bordon-Pallier, F., Bronstrup, M., Courtemanche, G., Gerlitz, M., Geslin, M., Hammann, P., Heinz, D.W., Hoffmann, H., Klieber, S., Kohlmann, M., Kurz, M., Lair, C., Matter, H., Nuermberger, E., Tyagi, S., Fraisse, L., Grosset, J.H., Lagrange, S., Muller, R.(2015) Science 348: 1106
- PubMed: 26045430
- DOI: https://doi.org/10.1126/science.aaa4690
- Primary Citation of Related Structures:
5AGU, 5AGV, 5AH2, 5AH4 - PubMed Abstract:
The discovery of Streptomyces-produced streptomycin founded the age of tuberculosis therapy. Despite the subsequent development of a curative regimen for this disease, tuberculosis remains a worldwide problem, and the emergence of multidrug-resistant Mycobacterium tuberculosis has prioritized the need for new drugs. Here we show that new optimized derivatives from Streptomyces-derived griselimycin are highly active against M. tuberculosis, both in vitro and in vivo, by inhibiting the DNA polymerase sliding clamp DnaN. We discovered that resistance to griselimycins, occurring at very low frequency, is associated with amplification of a chromosomal segment containing dnaN, as well as the ori site. Our results demonstrate that griselimycins have high translational potential for tuberculosis treatment, validate DnaN as an antimicrobial target, and capture the process of antibiotic pressure-induced gene amplification.
- Department of Microbial Natural Products, Helmholtz Institute for Pharmaceutical Research Saarland (HIPS), Helmholtz Centre for Infection Research and Pharmaceutical Biotechnology, Saarland University, 66123 Saarbrücken, Germany. German Centre for Infection Research (DZIF), Partner Site Hannover-Braunschweig, Hannover, Germany.
Organizational Affiliation: