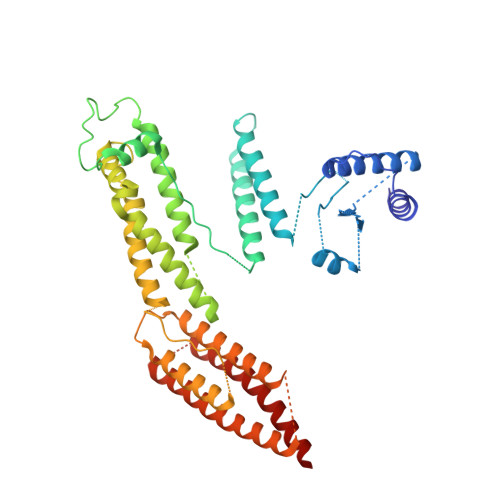
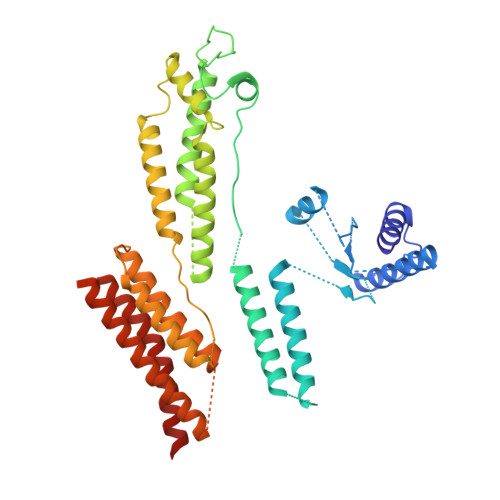
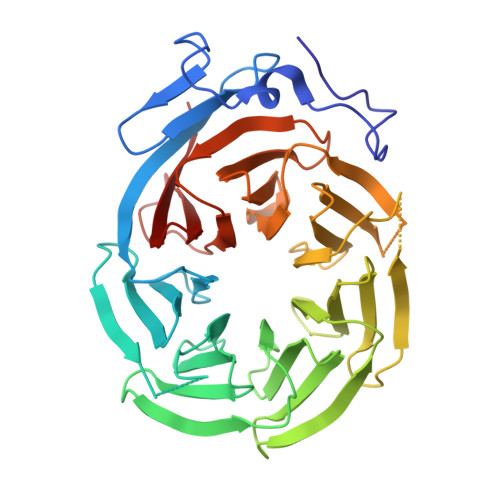
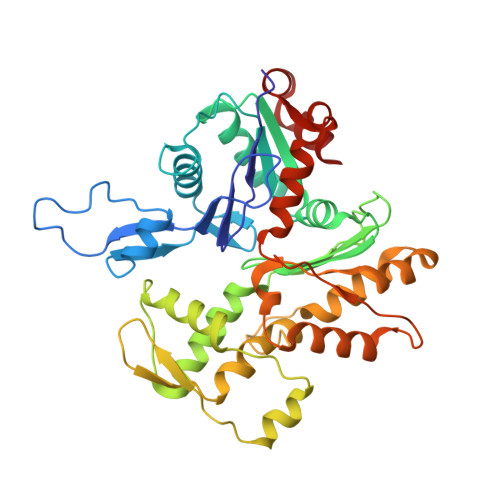
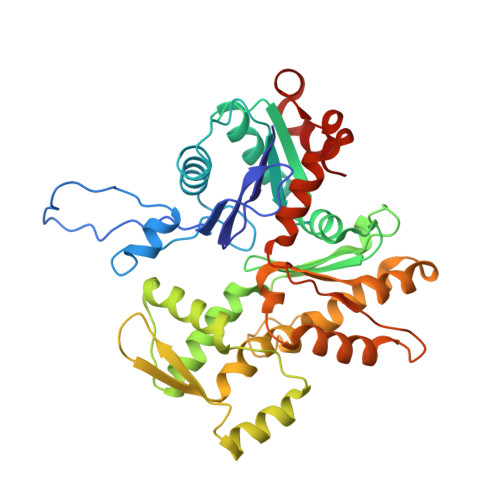
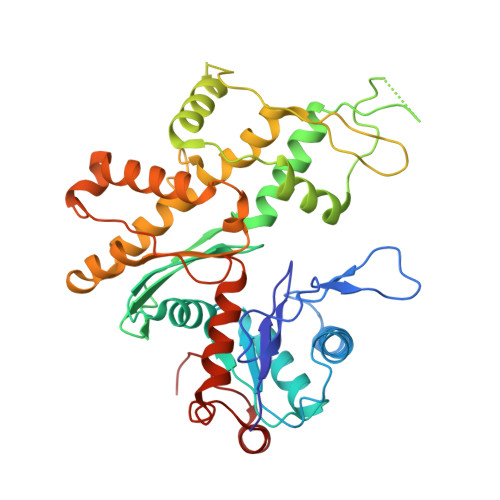
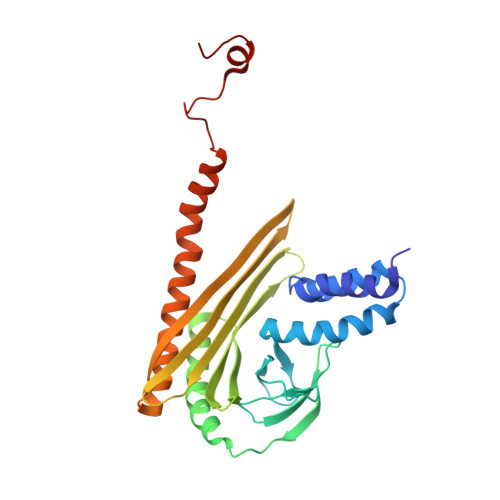
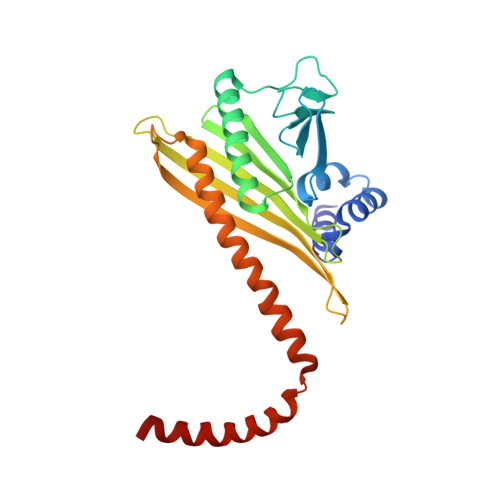
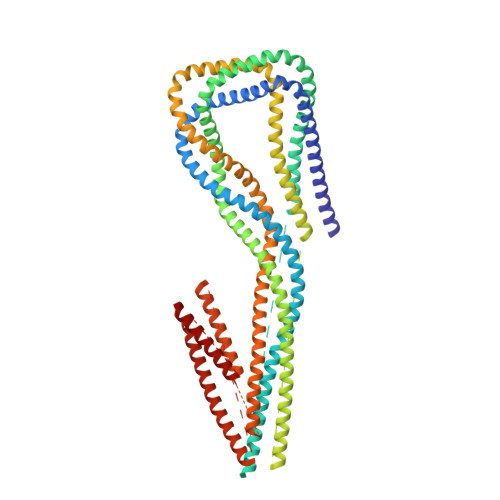
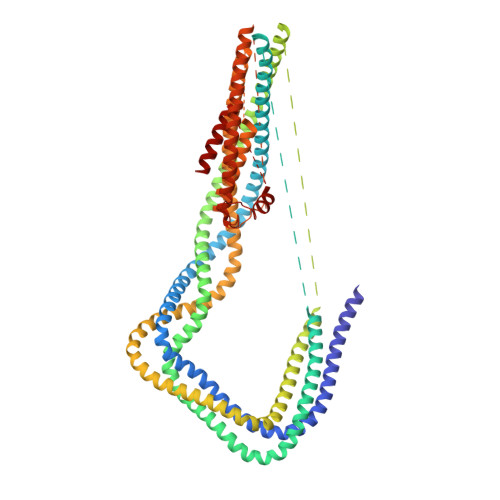
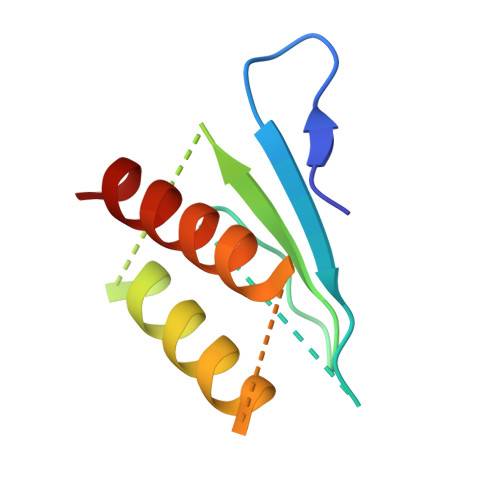
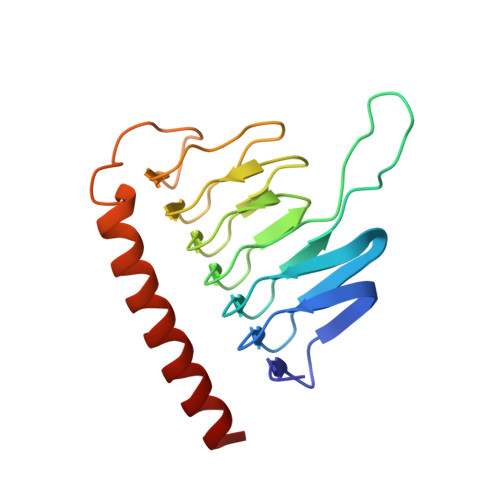
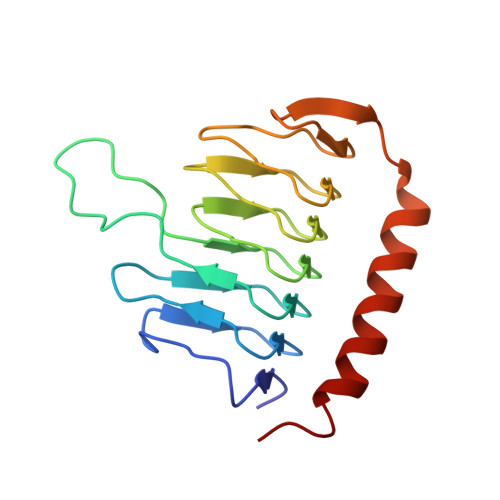
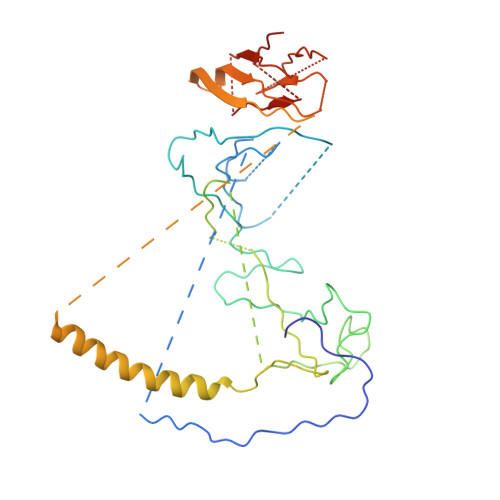
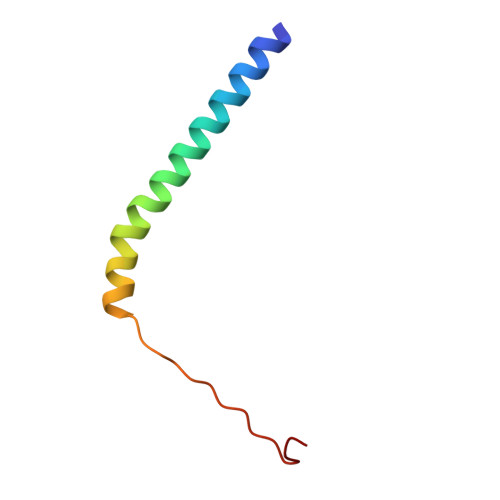
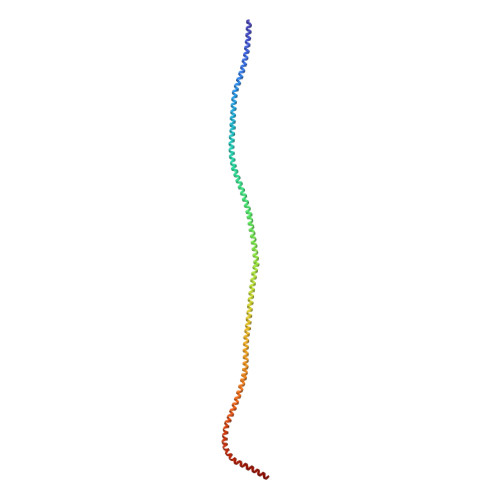The Structure of the Dynactin Complex and its Interaction with Dynein.
Urnavicius, L., Zhang, K., Diamant, A.G., Motz, C., Schlager, M.A., Yu, M., Patel, N.A., Robinson, C.V., Carter, A.P.(2015) Science 347: 1441
- PubMed: 25814576
- DOI: https://doi.org/10.1126/science.aaa4080
- Primary Citation of Related Structures:
5ADX, 5AFR, 5AFU - PubMed Abstract:
Dynactin is an essential cofactor for the microtubule motor cytoplasmic dynein-1. We report the structure of the 23-subunit dynactin complex by cryo-electron microscopy to 4.0 angstroms. Our reconstruction reveals how dynactin is built around a filament containing eight copies of the actin-related protein Arp1 and one of β-actin. The filament is capped at each end by distinct protein complexes, and its length is defined by elongated peptides that emerge from the α-helical shoulder domain. A further 8.2 angstrom structure of the complex between dynein, dynactin, and the motility-inducing cargo adaptor Bicaudal-D2 shows how the translational symmetry of the dynein tail matches that of the dynactin filament. The Bicaudal-D2 coiled coil runs between dynein and dynactin to stabilize the mutually dependent interactions between all three components.
- Medical Research Council Laboratory of Molecular Biology, Division of Structural Studies, Francis Crick Avenue, Cambridge, CB2 0QH, UK.
Organizational Affiliation: