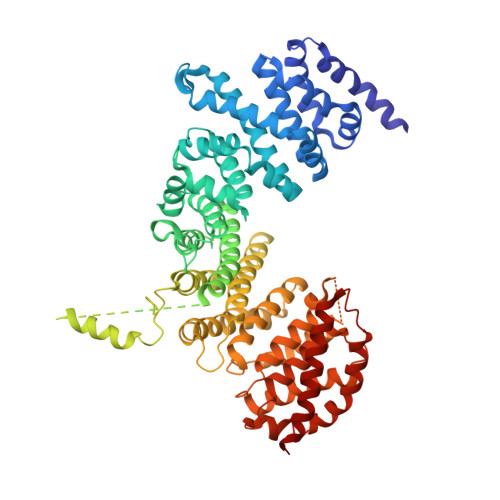
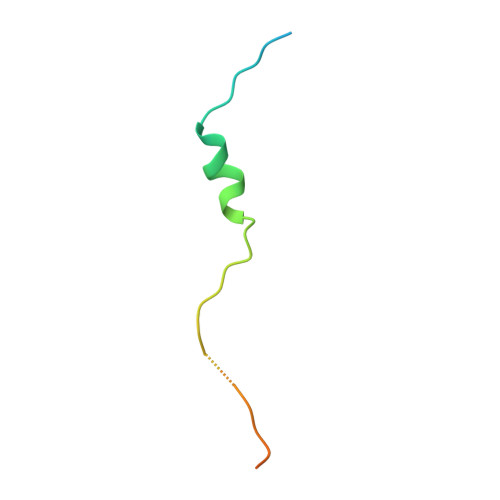
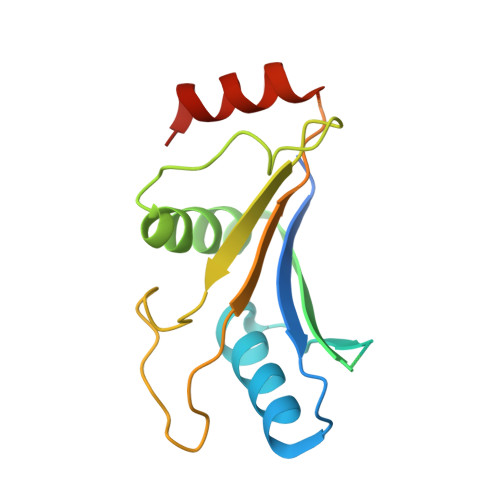
Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Calvino, F.R., Kharde, S., Ori, A., Hendricks, A., Wild, K., Kressler, D., Bange, G., Hurt, E., Beck, M., Sinning, I.(2015) Nat Commun 6: 6510
- PubMed: 25849277
- DOI: https://doi.org/10.1038/ncomms7510
- Primary Citation of Related Structures:
5AFF - PubMed Abstract:
During 60S biogenesis, mature 5S RNP consisting of 5S RNA, RpL5 and RpL11, assembles into a pre-60S particle, where docking relies on RpL11 interacting with helix 84 (H84) of the 25S RNA. How 5S RNP is assembled for recruitment into the pre-60S is not known. Here we report the crystal structure of a ternary symportin Syo1-RpL5-N-RpL11 complex and provide biochemical and structural insights into 5S RNP assembly. Syo1 guards the 25S RNA-binding surface on RpL11 and competes with H84 for binding. Pull-down experiments show that H84 releases RpL11 from the ternary complex, but not in the presence of 5S RNA. Crosslinking mass spectrometry visualizes structural rearrangements on incorporation of 5S RNA into the Syo1-RpL5-RpL11 complex supporting the formation of a pre-5S RNP. Our data underline the dual role of Syo1 in ribosomal protein transport and as an assembly platform for 5S RNP.
- Heidelberg University Biochemistry Center, Im Neuenheimer Feld 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: