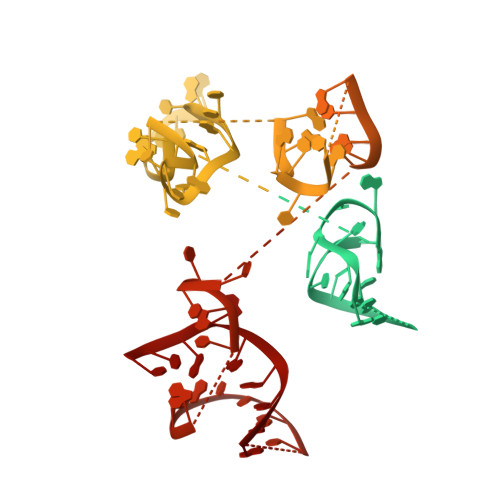
Structure of a Human Translation Termination Complex.
Matheisl, S., Berninghausen, O., Becker, T., Beckmann, R.(2015) Nucleic Acids Res 43: 8615
- PubMed: 26384426
- DOI: https://doi.org/10.1093/nar/gkv909
- Primary Citation of Related Structures:
5A8L - PubMed Abstract:
In contrast to bacteria that have two release factors, RF1 and RF2, eukaryotes only possess one unrelated release factor eRF1, which recognizes all three stop codons of the mRNA and hydrolyses the peptidyl-tRNA bond. While the molecular basis for bacterial termination has been elucidated, high-resolution structures of eukaryotic termination complexes have been lacking. Here we present a 3.8 Å structure of a human translation termination complex with eRF1 decoding a UAA(A) stop codon. The complex was formed using the human cytomegalovirus (hCMV) stalling peptide, which perturbs the peptidyltransferase center (PTC) to silence the hydrolysis activity of eRF1. Moreover, unlike sense codons or bacterial stop codons, the UAA stop codon adopts a U-turn-like conformation within a pocket formed by eRF1 and the ribosome. Inducing the U-turn conformation for stop codon recognition rationalizes how decoding by eRF1 includes monitoring geometry in order to discriminate against sense codons.
- Gene Center and Center for integrated Protein Science Munich, Department of Biochemistry, Feodor-Lynen-Str. 25, University of Munich, 81377 Munich, Germany.
Organizational Affiliation: