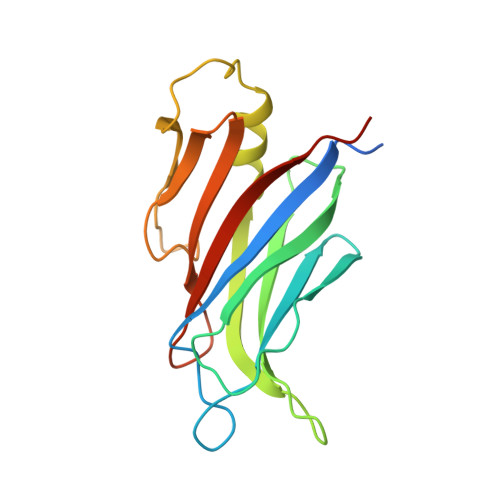
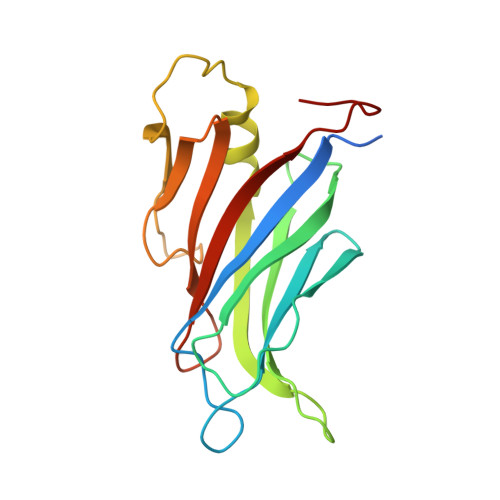
Calcium-Dependent Oligomerization of Car Proteins at Cell Membrane Modulates Aba Signaling.
Diaz, M., Sanchez-Barrena, M.J., Gonzalez-Rubio, J.M., Rodriguez, L., Fernandez, D., Antoni, R., Yunta, C., Belda-Palazon, B., Gonzalez-Guzman, M., Peirats-Llobet, M., Menendez, M., Boskovic, J., Marquez, J.A., Rodriguez, P.L., Albert, A.(2016) Proc Natl Acad Sci U S A 113: E396
- PubMed: 26719420
- DOI: https://doi.org/10.1073/pnas.1512779113
- Primary Citation of Related Structures:
5A4X, 5A50, 5A51, 5A52 - PubMed Abstract:
Regulation of ion transport in plants is essential for cell function. Abiotic stress unbalances cell ion homeostasis, and plants tend to readjust it, regulating membrane transporters and channels. The plant hormone abscisic acid (ABA) and the second messenger Ca(2+) are central in such processes, as they are involved in the regulation of protein kinases and phosphatases that control ion transport activity in response to environmental stimuli. The identification and characterization of the molecular mechanisms underlying the effect of ABA and Ca(2+) signaling pathways on membrane function are central and could provide opportunities for crop improvement. The C2-domain ABA-related (CAR) family of small proteins is involved in the Ca(2+)-dependent recruitment of the pyrabactin resistance 1/PYR1-like (PYR/PYL) ABA receptors to the membrane. However, to fully understand CAR function, it is necessary to define a molecular mechanism that integrates Ca(2+) sensing, membrane interaction, and the recognition of the PYR/PYL interacting partners. We present structural and biochemical data showing that CARs are peripheral membrane proteins that functionally cluster on the membrane and generate strong positive membrane curvature in a Ca(2+)-dependent manner. These features represent a mechanism for the generation, stabilization, and/or specific recognition of membrane discontinuities. Such structures may act as signaling platforms involved in the recruitment of PYR/PYL receptors and other signaling components involved in cell responses to stress.
- Instituto de Química Física Rocasolano, Consejo Superior de Investigaciones Científicas, ES-28006 Madrid, Spain;
Organizational Affiliation: