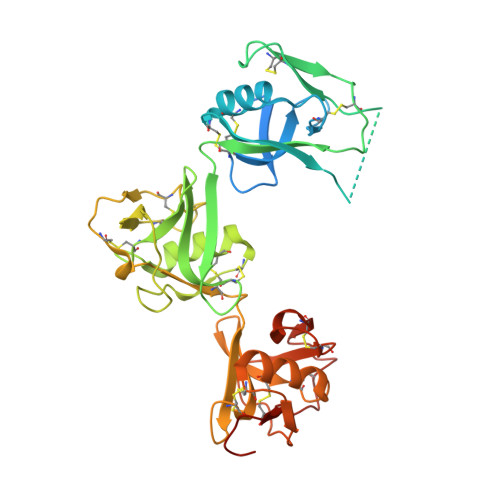
Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Chappell, P.E., Garner, L.I., Yan, J., Metcalfe, C., Hatherley, D., Johnson, S., Robinson, C.V., Lea, S.M., Brown, M.H.(2015) Structure 23: 1426
- PubMed: 26146185
- DOI: https://doi.org/10.1016/j.str.2015.05.019
- Primary Citation of Related Structures:
5A2E, 5A2F - PubMed Abstract:
CD6 is a transmembrane protein with an extracellular region containing three scavenger receptor cysteine rich (SRCR) domains. The membrane proximal domain of CD6 binds the N-terminal immunoglobulin superfamily (IgSF) domain of another cell surface receptor, CD166, which also engages in homophilic interactions. CD6 expression is mainly restricted to T cells, and the interaction between CD6 and CD166 regulates T-cell activation. We have solved the X-ray crystal structures of the three SRCR domains of CD6 and two N-terminal domains of CD166. This first structure of consecutive SRCR domains reveals a nonlinear organization. We characterized the binding sites on CD6 and CD166 and showed that a SNP in CD6 causes glycosylation that hinders the CD6/CD166 interaction. Native mass spectrometry analysis showed that there is competition between the heterophilic and homophilic interactions. These data give insight into how interactions of consecutive SRCR domains are perturbed by SNPs and potential therapeutic reagents.
- Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, UK.
Organizational Affiliation: