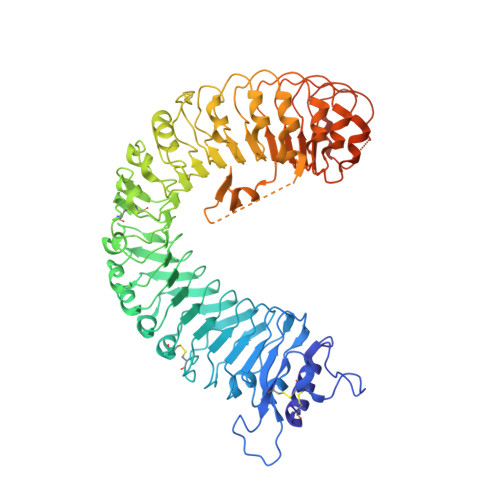
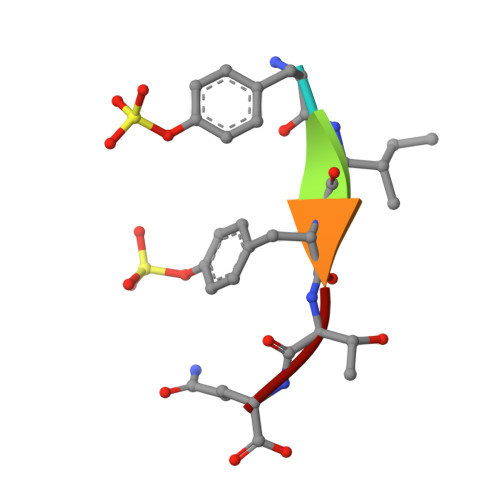
Allosteric receptor activation by the plant peptide hormone phytosulfokine
Wang, J., Li, H., Han, Z., Zhang, H., Wang, T., Lin, G., Chang, J., Yang, W., Chai, J.(2015) Nature 525: 265-268
- PubMed: 26308901
- DOI: https://doi.org/10.1038/nature14858
- Primary Citation of Related Structures:
4Z5W, 4Z61, 4Z62, 4Z63, 4Z64 - PubMed Abstract:
Phytosulfokine (PSK) is a disulfated pentapeptide that has a ubiquitous role in plant growth and development. PSK is perceived by its receptor PSKR, a leucine-rich repeat receptor kinase (LRR-RK). The mechanisms underlying the recognition of PSK, the activation of PSKR and the identity of the components downstream of the initial binding remain elusive. Here we report the crystal structures of the extracellular LRR domain of PSKR in free, PSK- and co-receptor-bound forms. The structures reveal that PSK interacts mainly with a β-strand from the island domain of PSKR, forming an anti-β-sheet. The two sulfate moieties of PSK interact directly with PSKR, sensitizing PSKR recognition of PSK. Supported by biochemical, structural and genetic evidence, PSK binding enhances PSKR heterodimerization with the somatic embryogenesis receptor-like kinases (SERKs). However, PSK is not directly involved in PSKR-SERK interaction but stabilizes PSKR island domain for recruitment of a SERK. Our data reveal the structural basis for PSKR recognition of PSK and allosteric activation of PSKR by PSK, opening up new avenues for the design of PSKR-specific small molecules.
- Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: